Exploring and mapping chemical space with molecular assembly trees
- PMID: 34559562
- PMCID: PMC8462901
- DOI: 10.1126/sciadv.abj2465
Exploring and mapping chemical space with molecular assembly trees
Abstract
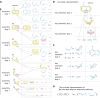
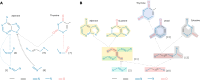
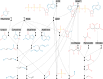
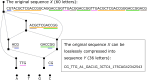
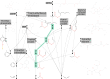
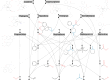
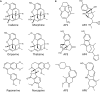
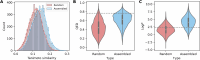
The rule-based search of chemical space can generate an almost infinite number of molecules, but exploration of known molecules as a function of the minimum number of steps needed to build up the target graphs promises to uncover new motifs and transformations. Assembly theory is an approach to compare the intrinsic complexity and properties of molecules by the minimum number of steps needed to build up the target graphs. Here, we apply this approach to prebiotic chemistry, gene sequences, plasticizers, and opiates. This allows us to explore molecules connected to the assembly tree, rather than the entire space of molecules possible. Last, by developing a reassembly method, based on assembly trees, we found that in the case of the opiates, a new set of drug candidates could be generated that would not be accessible via conventional fragment-based drug design, thereby demonstrating how this approach might find application in drug discovery.
Figures
References
-
- Dobson C. M., Chemical space and biology. Nature 432, 824–828 (2004). - PubMed
-
- Lipinski C., Hopkins A., Navigating chemical space for biology and medicine. Nature 432, 855–861 (2004). - PubMed
-
- Bohacek R. S., McMartin C., Guida W. C., The art and practice of structure-based drug design: A molecular modeling perspective. Med. Res. Rev. 16, 3–50 (1996). - PubMed
-
- Kirkpatrick P., Ellis C., Chemical space. Nature 432, 823–823 (2004).
LinkOut - more resources
Full Text Sources
Other Literature Sources

