Supernova: A Deoxyribozyme that Catalyzes a Chemiluminescent Reaction
- PMID: 34559935
- PMCID: PMC9298802
- DOI: 10.1002/anie.202109347
Supernova: A Deoxyribozyme that Catalyzes a Chemiluminescent Reaction
Abstract
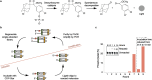
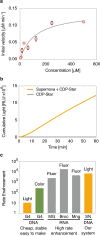
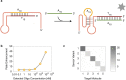
Functional DNA molecules are useful components in nanotechnology and synthetic biology. To expand the toolkit of functional DNA parts, in this study we used artificial evolution to identify a glowing deoxyribozyme called Supernova. This deoxyribozyme transfers a phosphate from a 1,2-dioxetane substrate to its 5' hydroxyl group, which triggers a chemiluminescent reaction and a flash of blue light. An engineered version of Supernova is only catalytically active in the presence of an oligonucleotide complementary to its 3' end, demonstrating that light production can be coupled to ligand binding. We anticipate that Supernova will be useful in a wide variety of applications, including as a signaling component in allosterically regulated sensors and in logic gates of molecular computers.
Keywords: aptazyme; catalytic DNA; chemiluminescence; deoxyribozyme; in vitro selection; luminescence; sensor.
© 2021 The Authors. Angewandte Chemie International Edition published by Wiley-VCH GmbH.
Conflict of interest statement
The authors have applied for a patent covering the use of this light‐producing deoxyribozyme Inventors: Katerina Svehlova, Ondřej Lukšan, Martin Jakubec, and Edward A. Curtis Application number: EP21156992.6 Status: pending.
Figures

References
-
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R., Cell 1982, 31, 147–157. - PubMed
-
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S., Cell 1983, 35, 849–857. - PubMed
-
- Tuerk C., Gold L., Science 1990, 249, 505–510. - PubMed
-
- Robertson D. L., Joyce G. F., Nature 1990, 344, 467–468. - PubMed
-
- Ellington A. D., Szostak J. W., Nature 1990, 346, 818–822. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

