Mitochondrial respiratory chain function promotes extracellular matrix integrity in cartilage
- PMID: 34560099
- PMCID: PMC8503590
- DOI: 10.1016/j.jbc.2021.101224
Mitochondrial respiratory chain function promotes extracellular matrix integrity in cartilage
Abstract
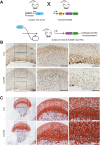
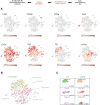
Energy metabolism and extracellular matrix (ECM) function together orchestrate and maintain tissue organization, but crosstalk between these processes is poorly understood. Here, we used single-cell RNA-Seq (scRNA-Seq) analysis to uncover the importance of the mitochondrial respiratory chain for ECM homeostasis in mature cartilage. This tissue produces large amounts of a specialized ECM to promote skeletal growth during development and maintain mobility throughout life. A combined approach of high-resolution scRNA-Seq, mass spectrometry/matrisome analysis, and atomic force microscopy was applied to mutant mice with cartilage-specific inactivation of respiratory chain function. This genetic inhibition in cartilage results in the expansion of a central area of 1-month-old mouse femur head cartilage, showing disorganized chondrocytes and increased deposition of ECM material. scRNA-Seq analysis identified a cell cluster-specific decrease in mitochondrial DNA-encoded respiratory chain genes and a unique regulation of ECM-related genes in nonarticular chondrocytes. These changes were associated with alterations in ECM composition, a shift in collagen/noncollagen protein content, and an increase of collagen crosslinking and ECM stiffness. These results demonstrate that mitochondrial respiratory chain dysfunction is a key factor that can promote ECM integrity and mechanostability in cartilage and presumably also in many other tissues.
Keywords: MMP10; THBS1; atomic force microscopy; extracellular matrix; matrisome; matrix metalloproteinase; mitochondria; mitochondrial respiratory chain; single-cell RNA-Seq; transcriptomics.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Romani P., Valcarcel-Jimenez L., Frezza C., Dupont S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 2021;22:22–38. - PubMed
-
- Weiland D., Brachvogel B., Hornig-Do H.T., Neuhaus J.F.G., Holzer T., Tobin D.J., Niessen C.M., Wiesner R.J., Baris O.R. Imbalance of mitochondrial respiratory chain complexes in the epidermis induces severe skin inflammation. J. Invest. Dermatol. 2018;138:132–140. - PubMed
-
- Baris O.R., Ederer S., Neuhaus J.F., von Kleist-Retzow J.C., Wunderlich C.M., Pal M., Wunderlich F.T., Peeva V., Zsurka G., Kunz W.S., Hickethier T., Bunck A.C., Stockigt F., Schrickel J.W., Wiesner R.J. Mosaic deficiency in mitochondrial oxidative metabolism promotes cardiac arrhythmia during aging. Cell Metab. 2015;21:667–677. - PubMed
-
- Ovchinnikov D.A., Deng J.M., Ogunrinu G., Behringer R.R. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. - PubMed
-
- Bluhm B., Ehlen H.W.A., Holzer T., Georgieva V.S., Heilig J., Pitzler L., Etich J., Bortecen T., Frie C., Probst K., Niehoff A., Belluoccio D., Van den Bergen J., Brachvogel B. miR-322 stabilizes MEK1 expression to inhibit RAF/MEK/ERK pathway activation in cartilage. Development. 2017;144:3562–3577. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

