A standardized analytics pipeline for reliable and rapid development and validation of prediction models using observational health data
- PMID: 34560604
- PMCID: PMC8420135
- DOI: 10.1016/j.cmpb.2021.106394
A standardized analytics pipeline for reliable and rapid development and validation of prediction models using observational health data
Abstract
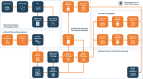
Background and objective: As a response to the ongoing COVID-19 pandemic, several prediction models in the existing literature were rapidly developed, with the aim of providing evidence-based guidance. However, none of these COVID-19 prediction models have been found to be reliable. Models are commonly assessed to have a risk of bias, often due to insufficient reporting, use of non-representative data, and lack of large-scale external validation. In this paper, we present the Observational Health Data Sciences and Informatics (OHDSI) analytics pipeline for patient-level prediction modeling as a standardized approach for rapid yet reliable development and validation of prediction models. We demonstrate how our analytics pipeline and open-source software tools can be used to answer important prediction questions while limiting potential causes of bias (e.g., by validating phenotypes, specifying the target population, performing large-scale external validation, and publicly providing all analytical source code).
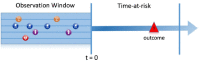
Methods: We show step-by-step how to implement the analytics pipeline for the question: 'In patients hospitalized with COVID-19, what is the risk of death 0 to 30 days after hospitalization?'. We develop models using six different machine learning methods in a USA claims database containing over 20,000 COVID-19 hospitalizations and externally validate the models using data containing over 45,000 COVID-19 hospitalizations from South Korea, Spain, and the USA.
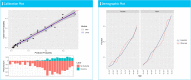
Results: Our open-source software tools enabled us to efficiently go end-to-end from problem design to reliable Model Development and evaluation. When predicting death in patients hospitalized with COVID-19, AdaBoost, random forest, gradient boosting machine, and decision tree yielded similar or lower internal and external validation discrimination performance compared to L1-regularized logistic regression, whereas the MLP neural network consistently resulted in lower discrimination. L1-regularized logistic regression models were well calibrated.
Conclusion: Our results show that following the OHDSI analytics pipeline for patient-level prediction modelling can enable the rapid development towards reliable prediction models. The OHDSI software tools and pipeline are open source and available to researchers from all around the world.
Keywords: COVID-19; Data harmonization; Data quality control; Distributed data network; Machine learning; Risk prediction.
Copyright © 2021. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest CB, MJS, AGS, JMR are employees of Janssen Research & Development and shareholders of Johnson & Johnson.
Figures




References
-
- World Health Organization . COVID-19 weekly epidemiological update, edition 45, 22 June 2021. World Health Organization [Online]; 2021. https://apps.who.int/iris/handle/10665/342009 [Online]. Available:
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

