TGFBI Production by Macrophages Contributes to an Immunosuppressive Microenvironment in Ovarian Cancer
- PMID: 34561272
- PMCID: PMC9397609
- DOI: 10.1158/0008-5472.CAN-21-0536
TGFBI Production by Macrophages Contributes to an Immunosuppressive Microenvironment in Ovarian Cancer
Abstract
The tumor microenvironment evolves during malignant progression, with major changes in nonmalignant cells, cytokine networks, and the extracellular matrix (ECM). In this study, we aimed to understand how the ECM changes during neoplastic transformation of serous tubal intraepithelial carcinoma lesions (STIC) into high-grade serous ovarian cancers (HGSOC). Analysis of the mechanical properties of human fallopian tubes (FT) and ovaries revealed that normal FT and fimbria had a lower tissue modulus, a measure of stiffness, than normal or diseased ovaries. Proteomic analysis of the matrisome fraction between FT, fimbria, and ovaries showed significant differences in the ECM protein TGF beta induced (TGFBI, also known as βig-h3). STIC lesions in the fimbria expressed high levels of TGFBI, which was predominantly produced by CD163-positive macrophages proximal to STIC epithelial cells. In vitro stimulation of macrophages with TGFβ and IL4 induced secretion of TGFBI, whereas IFNγ/LPS downregulated macrophage TGFBI expression. Immortalized FT secretory epithelial cells carrying clinically relevant TP53 mutations stimulated macrophages to secrete TGFBI and upregulated integrin αvβ3, a putative TGFBI receptor. Transcriptomic HGSOC datasets showed a significant correlation between TGFBI expression and alternatively activated macrophage signatures. Fibroblasts in HGSOC metastases expressed TGFBI and stimulated macrophage TGFBI production in vitro. Treatment of orthotopic mouse HGSOC tumors with an anti-TGFBI antibody reduced peritoneal tumor size, increased tumor monocytes, and activated β3-expressing unconventional T cells. In conclusion, TGFBI may favor an immunosuppressive microenvironment in STICs that persists in advanced HGSOC. Furthermore, TGFBI may be an effector of the tumor-promoting actions of TGFβ and a potential therapeutic target. SIGNIFICANCE: Analysis of ECM changes during neoplastic transformation reveals a role for TGFBI secreted by macrophages in immunosuppression in early ovarian cancer.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures


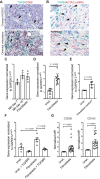
![Figure 4. Stimulation of TGFBI expression in macrophages by p53 mutated FTSE cells resembling STICs. A, Expression of p53 in STIC lesions. B, Expression of p53 in FT318 wild-type and mutant p53 transduced FTSE cells (R175H, R273H, R273C, R248W, Y220C). β-actin was used as loading control. C, TGFBI transcript expression of unstimulated and TGFβ-stimulated FT318 wild-type and mutant p53 FTSE cell lines in comparison with unstimulated (unst.) and TGFβ-stimulated macrophages. Data (n = 3) shown are mean ± SEM. Statistical significance was determined using one-way ANOVA with Dunnett multiple comparisons test. D, TGFBI expression in macrophages cultured in transwells with FTSE wild-type and FTSE-mutant cell lines. Data are shown for unstimulated macrophages (n = 7), FT318 (n = 3), R175H (n = 3), R273H (n = 3), R273C (n = 3), R248W (n = 3), and Y220C (n = 3). Statistical significance was determined using one-way ANOVA with Dunnett multiple comparisons test.E, TGFBI secretion of macrophages alone (M0) or cocultured with FTSE cell lines [wild type (FNE01, FNE02, FT318) and mutant p53 (R175H, R273H, R273C, R248W, Y220C)] and a HGSOC cell line (G164) normalized for 106 macrophages. Data are mean ± SD. Macrophages (n = 7), FNE01 (n = 6), FNE02 (n = 6), FT318 (n = 7), R175H (n = 4), R273H (n = 4), R273C (n = 4), R248W (n = 4), Y220C (n = 4), G164 (n = 6). Statistical significance determined using one-way ANOVA with Dunnett multiple comparisons test. Compared with coculture with FT318, secretion of TGFBI is significant only for cell lines R273C and Y220C. F, TGFBI secretion of cocultured macrophages with FTSE and HGSOC cells in the presence or absence of the selective TGFβR inhibitor SB431542. Data are mean ± SD. n = 3 for all the inhibitor-treated conditions. Statistical significance was determined using one-way ANOVA with Dunnett multiple comparisons test. Untreated coculture data points are the same as in E. G, IHC staining of healthy and diseased human tissues for the integrin subunits αv and β3. Representative data are shown for FT (n = 3), FB (n = 3), STICs (n = 7), ovary (n = 3), and HGSOC diseased ovary (n = 4) tissues. H, FT318 wild-type and mutant p53 (R175H, R273H, R273C, R248W, Y220C) FTSE cells stained for the integrin αvβ3 [n = 5 (FT318, R273C, R273H); n = 3 (R175H, R248W, Y220C)] and the subunit β3 (n = 3). Data are mean ± SEM. Statistical significance determined using one-way ANOVA with Bonferroni multiple comparisons test. MFI, mean fluorescence intensity.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/882c/9397609/0c3704661587/5706fig4.gif)


References
-
- Mehra K, Mehrad M, Ning G, Drapkin R, McKeon FD, Xian W, et al. . STICS, SCOUTs and p53 signatures; a new language for pelvic serous carcinogenesis. Front Biosci 2011;3:625–34. - PubMed
-
- Lee Y, Miron A, Drapkin R, Nucci MR, Medeiros F, Saleemuddin A, et al. . A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J Pathol 2007;211:26–35. - PubMed
-
- Kindelberger DW, Lee Y, Miron A, Hirsch MS, Feltmate C, Medeiros F, et al. . Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: evidence for a causal relationship. Am J Surg Pathol 2007;31:161–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 27947/CRUK_/Cancer Research UK/United Kingdom
- C16420/A18066/CRUK_/Cancer Research UK/United Kingdom
- A21222/CRUK_/Cancer Research UK/United Kingdom
- 16354/CRUK_/Cancer Research UK/United Kingdom
- P50 CA228991/CA/NCI NIH HHS/United States
- 15601/CRUK_/Cancer Research UK/United Kingdom
- C587/A25714/CRUK_/Cancer Research UK/United Kingdom
- C587/A16354/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 25714/CRUK_/Cancer Research UK/United Kingdom
- C355/A25137/CRUK_/Cancer Research UK/United Kingdom
- 22905/CRUK_/Cancer Research UK/United Kingdom
- 201118/Z/16/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

