Protease activity sensors enable real-time treatment response monitoring in lymphangioleiomyomatosis
- PMID: 34561286
- PMCID: PMC9030069
- DOI: 10.1183/13993003.00664-2021
Protease activity sensors enable real-time treatment response monitoring in lymphangioleiomyomatosis
Abstract
Background: Biomarkers of disease progression and treatment response are urgently needed for patients with lymphangioleiomyomatosis (LAM). Activity-based nanosensors, an emerging biosensor class, detect dysregulated proteases in vivo and release a reporter to provide a urinary readout of disease. Because proteases are dysregulated in LAM and may directly contribute to lung function decline, activity-based nanosensors may enable quantitative, real-time monitoring of LAM progression and treatment response. We aimed to assess the diagnostic utility of activity-based nanosensors in a pre-clinical model of pulmonary LAM.
Methods: Tsc2-null cells were injected intravenously into female nude mice to establish a mouse model of pulmonary LAM. A library of 14 activity-based nanosensors, designed to detect proteases across multiple catalytic classes, was administered into the lungs of LAM mice and healthy controls, urine was collected, and mass spectrometry was performed to measure nanosensor cleavage products. Mice were then treated with rapamycin and monitored with activity-based nanosensors. Machine learning was performed to distinguish diseased from healthy and treated from untreated mice.
Results: Multiple activity-based nanosensors (PP03 (cleaved by metallo, aspartic and cysteine proteases), padjusted<0.0001; PP10 (cleaved by serine, aspartic and cysteine proteases), padjusted=0.017)) were differentially cleaved in diseased and healthy lungs, enabling strong classification with a machine learning model (area under the curve (AUC) 0.95 from healthy). Within 2 days after rapamycin initiation, we observed normalisation of PP03 and PP10 cleavage, and machine learning enabled accurate classification of treatment response (AUC 0.94 from untreated).
Conclusions: Activity-based nanosensors enable noninvasive, real-time monitoring of disease burden and treatment response in a pre-clinical model of LAM.
Copyright ©The authors 2022.
Conflict of interest statement
Conflict of interest: J.D. Kirkpatrick reports grants from the Ludwig Fund for Cancer Research, during the conduct of the study. In addition, J.D. Kirkpatrick has a patent pending (Lung protease nanosensors and uses thereof; PCT/US2019/052868, filed 25 September 2019). Conflict of interest: A.P. Soleimany has nothing to disclose. Conflict of interest: J.S. Dudani has nothing to disclose. Conflict of interest: H-J. Liu has nothing to disclose. Conflict of interest: H.C. Lam has nothing to disclose. Conflict of interest: C. Priolo has nothing to disclose. Conflict of interest: E.P. Henske has nothing to disclose. Conflict of interest: S.N. Bhatia reports other support from the Howard Hughes Medical Institute and National Institute of Environmental Health Sciences, grants from the Ludwig Fund for Cancer Research and grants from the Koch Institute Marble Center for Cancer Nanomedicine, during the conduct of the study; other support from Vertex Pharmaceuticals, Glympse Bio, Maverick Therapeutics, Satellite Bio, CEND Rx and Moderna Therapeutics, outside the submitted work. In addition, S.N. Bhatia has a patent pending (Lung protease nanosensors and uses thereof; PCT/US2019/052868, filed 25 September 2019).
Figures
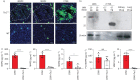
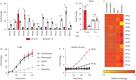


Comment in
-
Proteinases in the pathogenesis of lymphangioleiomyomatosis lung disease: nibbling or chewing up the lung?Eur Respir J. 2022 Apr 14;59(4):2200405. doi: 10.1183/13993003.00405-2022. Print 2022 Apr. Eur Respir J. 2022. PMID: 35422429 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
