A longitudinal sampling study of transcriptomic and epigenetic profiles in patients with thrombocytopenia syndrome
- PMID: 34561445
- PMCID: PMC8463551
- DOI: 10.1038/s41467-021-25804-z
A longitudinal sampling study of transcriptomic and epigenetic profiles in patients with thrombocytopenia syndrome
Abstract
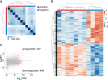
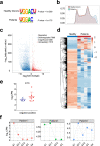
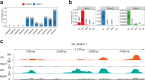
Severe fever with thrombocytopenia syndrome (SFTS) is a novel tick-borne infectious disease caused by a new type of SFTS virus (SFTSV). Here, a longitudinal sampling study is conducted to explore the differences in transcript levels after SFTSV infection, and to characterize the transcriptomic and epigenetic profiles of hospitalized patients. The results reveal significant changes in the mRNA expression of certain genes from onset to recovery. Moreover, m6A-seq reveals that certain genes related with immune regulation may be regulated by m6A. Besides the routine tests such as platelet counts, serum ALT and AST levels testing, distinct changes in myocardial enzymes, coagulation function, and inflammation are well correlated with the clinical data and sequencing data, suggesting that clinical practitioners should monitor the above indicators to track disease progression and guide personalized treatment. In this study, the transcript changes and RNA modification may lend a fresh perspective to our understanding of the SFTSV and play a significant role in the discovery of drugs for effective treatment of this disease.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

