Copper nitrite reductase from Sinorhizobium meliloti 2011: Crystal structure and interaction with the physiological versus a nonmetabolically related cupredoxin-like mediator
- PMID: 34562300
- PMCID: PMC8521279
- DOI: 10.1002/pro.4195
Copper nitrite reductase from Sinorhizobium meliloti 2011: Crystal structure and interaction with the physiological versus a nonmetabolically related cupredoxin-like mediator
Abstract
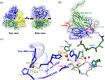
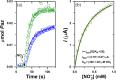
We report the crystal structure of the copper-containing nitrite reductase (NirK) from the Gram-negative bacterium Sinorhizobium meliloti 2011 (Sm), together with complex structural alignment and docking studies with both non-cognate and the physiologically related pseudoazurins, SmPaz1 and SmPaz2, respectively. S. meliloti is a rhizobacterium used for the formulation of Medicago sativa bionoculants, and SmNirK plays a key role in this symbiosis through the denitrification pathway. The structure of SmNirK, solved at a resolution of 2.5 Å, showed a striking resemblance with the overall structure of the well-known Class I NirKs composed of two Greek key β-barrel domains. The activity of SmNirK is ~12% of the activity reported for classical NirKs, which could be attributed to several factors such as subtle structural differences in the secondary proton channel, solvent accessibility of the substrate channel, and that the denitrifying activity has to be finely regulated within the endosymbiont. In vitro kinetics performed in homogenous and heterogeneous media showed that both SmPaz1 and SmPaz2, which are coded in different regions of the genome, donate electrons to SmNirK with similar performance. Even though the energetics of the interprotein electron transfer (ET) process is not favorable with either electron donors, adduct formation mediated by conserved residues allows minimizing the distance between the copper centers involved in the interprotein ET process.
Keywords: NirK; Sinorhizobium meliloti 2011; X-ray crystal structure; copper; nitrite reductase; pseudoazurin.
© 2021 The Protein Society.
Conflict of interest statement
All authors declare no conflict of interest.
Figures



References
-
- Jaiswal SK, Mohammed M, Ibny FYI, Dakora FD. Rhizobia as a source of plant growth‐promoting molecules: Potential applications and possible operational mechanisms. Front Sustain Food Syst. 2021;4:619676.
-
- Delgado MJ, Casella S, Bedmar EJ. Chapter 6—Denitrification in rhizobia‐legume Symbiosis. In: Bothe H, Ferguson SJ, Newton WE, editors. Biology of the nitrogen cycle. Amsterdam: Elsevier, 2007; p. 83–91.
-
- Gruber N, Galloway JN. An earth‐system perspective of the global nitrogen cycle. Nature. 2008;451:293–296. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

