A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis
- PMID: 34564416
- PMCID: PMC8469675
- DOI: 10.3390/metabo11090601
A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis
Abstract
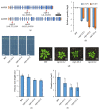
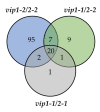
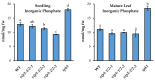
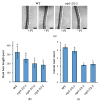
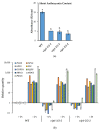
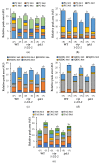
Phosphate is a major plant macronutrient and low phosphate availability severely limits global crop productivity. In Arabidopsis, a key regulator of the transcriptional response to low phosphate, phosphate starvation response 1 (PHR1), is modulated by a class of signaling molecules called inositol pyrophosphates (PP-InsPs). Two closely related diphosphoinositol pentakisphosphate enzymes (AtVIP1 and AtVIP2) are responsible for the synthesis and turnover of InsP8, the most implicated molecule. This study is focused on characterizing Arabidopsis vip1/vip2 double mutants and their response to low phosphate. We present evidence that both local and systemic responses to phosphate limitation are dampened in the vip1/vip2 mutants as compared to wild-type plants. Specifically, we demonstrate that under Pi-limiting conditions, the vip1/vip2 mutants have shorter root hairs and lateral roots, less accumulation of anthocyanin and less accumulation of sulfolipids and galactolipids. However, phosphate starvation response (PSR) gene expression is unaffected. Interestingly, many of these phenotypes are opposite to those exhibited by other mutants with defects in the PP-InsP synthesis pathway. Our results provide insight on the nexus between inositol phosphates and pyrophosphates involved in complex regulatory mechanisms underpinning phosphate homeostasis in plants.
Keywords: AtVIP1; AtVIP2; inositol pyrophosphates; lipid remodeling; phosphate homeostasis; phosphate starvation response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

