Establishment of a Newborn Lamb Gut-Loop Model to Evaluate New Methods of Enteric Disease Control and Reduce Experimental Animal Use
- PMID: 34564564
- PMCID: PMC8472880
- DOI: 10.3390/vetsci8090170
Establishment of a Newborn Lamb Gut-Loop Model to Evaluate New Methods of Enteric Disease Control and Reduce Experimental Animal Use
Abstract
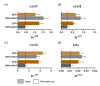
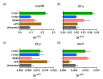
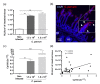
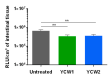
Enteric infectious diseases are not all well controlled, which leads to animal suffering and sometimes death in the most severe cases, in addition to economic losses for farmers. Typical symptoms of enteric infections include watery diarrhea, stomach cramps or pain, dehydration, nausea, vomiting, fever and weight loss. Evaluation of new control methods against enteric infections requires the use of many animals. We aimed to develop a new method for an initial in vivo screen of promising compounds against neonatal diseases such as cryptosporidiosis while limiting experimental animal use. We therefore adapted an in vivo method of multiple consecutive but independent intestinal loops to newborn lambs delivered by cesarean section, in which endotoxin responsiveness is retained. This new method allowed for the screening of natural yeast fractions for their ability to stimulate immune responses and to limit early Cryptosporidium parvum development. This model may also be used to investigate host-pathogen interactions and immune responses in a neonatal controlled environment.
Keywords: 3R; Cryptosporidium parvum; multiple intestinal loop model; newborn lamb; yeast cell wall fractions.
Conflict of interest statement
The authors declared no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
Figures

Similar articles
-
Genotyping and Subtyping Cryptosporidium To Identify Risk Factors and Transmission Patterns - Nebraska, 2015-2017.MMWR Morb Mortal Wkly Rep. 2020 Mar 27;69(12):335-338. doi: 10.15585/mmwr.mm6912a4. MMWR Morb Mortal Wkly Rep. 2020. PMID: 32214081 Free PMC article.
-
Innate immune responses play a key role in controlling infection of the intestinal epithelium by Cryptosporidium.Int J Parasitol. 2017 Oct;47(12):711-721. doi: 10.1016/j.ijpara.2017.08.001. Epub 2017 Sep 8. Int J Parasitol. 2017. PMID: 28893638 Review.
-
First report on Cryptosporidium parvum, Escherichia coli K99, rotavirus and coronavirus in neonatal lambs from north-center region, Algeria.Comp Immunol Microbiol Infect Dis. 2020 Dec;73:101567. doi: 10.1016/j.cimid.2020.101567. Epub 2020 Oct 22. Comp Immunol Microbiol Infect Dis. 2020. PMID: 33157428 Free PMC article.
-
Efficacy of chitosan, a natural polysaccharide, against Cryptosporidium parvum in vitro and in vivo in neonatal mice.Exp Parasitol. 2018 Nov;194:1-8. doi: 10.1016/j.exppara.2018.09.003. Epub 2018 Sep 17. Exp Parasitol. 2018. PMID: 30237052
-
Bovine cryptosporidiosis: impact, host-parasite interaction and control strategies.Vet Res. 2017 Aug 11;48(1):42. doi: 10.1186/s13567-017-0447-0. Vet Res. 2017. PMID: 28800747 Free PMC article. Review.
Cited by
-
Editorial: Special Issue "Addressing New Therapeutic Strategies Using Models".Vet Sci. 2023 Mar 17;10(3):230. doi: 10.3390/vetsci10030230. Vet Sci. 2023. PMID: 36977269 Free PMC article.
-
Characterization of intestinal mononuclear phagocyte subsets in young ruminants at homeostasis and during Cryptosporidium parvum infection.Front Immunol. 2024 May 2;15:1379798. doi: 10.3389/fimmu.2024.1379798. eCollection 2024. Front Immunol. 2024. PMID: 38756777 Free PMC article.
References
-
- Richmond J. The 3Rs—Past, present and future. Scand. J. Lab. Anim. Sci. 2000;27:84–92.
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

