Update of the Planktonic Diatom Genus Pseudo-nitzschia in Aotearoa New Zealand Coastal Waters: Genetic Diversity and Toxin Production
- PMID: 34564641
- PMCID: PMC8473122
- DOI: 10.3390/toxins13090637
Update of the Planktonic Diatom Genus Pseudo-nitzschia in Aotearoa New Zealand Coastal Waters: Genetic Diversity and Toxin Production
Abstract
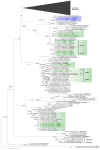
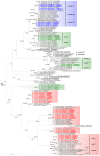
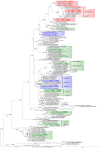
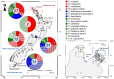
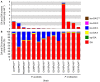
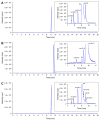
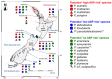
Domoic acid (DA) is produced by almost half of the species belonging to the diatom genus Pseudo-nitzschia and causes amnesic shellfish poisoning (ASP). It is, therefore, important to investigate the diversity and toxin production of Pseudo-nitzschia species for ASP risk assessments. Between 2018 and 2020, seawater samples were collected from various sites around Aotearoa New Zealand, and 130 clonal isolates of Pseudo-nitzschia were established. Molecular phylogenetic analysis of partial large subunit ribosomal DNA and/or internal transcribed spacer regions revealed that the isolates were divided into 14 species (Pseudo-nitzschia americana, Pseudo-nitzschia arenysensis, Pseudo-nitzschia australis, Pseudo-nitzschia calliantha, Pseudo-nitzschia cuspidata, Pseudo-nitzschia delicatissima, Pseudo-nitzschia fraudulenta, Pseudo-nitzschia galaxiae, Pseudo-nitzschia hasleana, Pseudo-nitzschia multiseries, Pseudo-nitzschia multistriata, Pseudo-nitzschia plurisecta, Pseudo-nitzschia pungens, and Pseudo-nitzschia cf. subpacifica). The P. delicatissima and P. hasleana strains were further divided into two clades/subclades (I and II). Liquid chromatography-tandem mass spectrometry was used to assess the production of DA and DA isomers by 73 representative strains. The analyses revealed that two (P. australis and P. multiseries) of the 14 species produced DA as a primary analogue, along with several DA isomers. This study is the first geographical distribution record of P. arenysensis, P.cuspidata, P. galaxiae, and P. hasleana in New Zealand coastal waters.
Keywords: Pacific Ocean; amnesic shellfish poisoning; domoic acid; geographical distribution; liquid chromatography-tandem mass spectrometry; molecular phylogenetic analysis; phytoplankton.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Subba Rao D.V., Quilliam M.A., Pocklington R. Domoic Acid—A Neurotoxic Amino Acid Produced by the Marine Diatom Nitzschia pungens in Culture. Can. J. Fish. Aquat Sci. 1988;45:2076–2079. doi: 10.1139/f88-241. - DOI
-
- Bates S.S., Bird C.J., de Freitas A.S.W., Foxall R., Gilgan M., Hanic L.A., Johnson G.R., McCulloch A.W., Odense P., Pocklington R., et al. Pennate Diatom Nitzschia pungens as the Primary Source of Domoic Acid, a Toxin in Shellfish from Eastern Prince Edward Island, Canada. Can. J. Fish. Aquat Sci. 1989;46:1203–1215. doi: 10.1139/f89-156. - DOI
-
- Hasle G.R. Pseudo-nitzschia pungens and P. multiseries (Bacillariophyceae): Nomenclatural History, Morphology, and Distribution. J. Phycol. 1995;31:428–435. doi: 10.1111/j.0022-3646.1995.00428.x. - DOI
-
- Hay B.E., Grant C.M., McCoubrey D.-J. A Review of the Marine Biotoxin Monitoring Programme for Non-Commercially Harvested Shellfish. NZ Ministry of Health; Wellington, New Zealand: 2000. Part 1: Technical report, A report prepared for the Ministry of Health by AquaBio consultants Ltd.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

