Clinical and functional characterization of a novel STUB1 frameshift mutation in autosomal dominant spinocerebellar ataxia type 48 (SCA48)
- PMID: 34565360
- PMCID: PMC8466936
- DOI: 10.1186/s12929-021-00763-1
Clinical and functional characterization of a novel STUB1 frameshift mutation in autosomal dominant spinocerebellar ataxia type 48 (SCA48)
Abstract
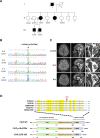
Background: Heterozygous pathogenic variants in STUB1 are implicated in autosomal dominant spinocerebellar ataxia type 48 (SCA48), which is a rare familial ataxia disorder. We investigated the clinical, genetic and functional characteristics of STUB1 mutations identified from a Taiwanese ataxia cohort.
Methods: We performed whole genome sequencing in a genetically undiagnosed family with an autosomal dominant ataxia syndrome. Further Sanger sequencing of all exons and intron-exon boundary junctions of STUB1 in 249 unrelated patients with cerebellar ataxia was performed. The pathogenicity of the identified novel STUB1 variant was investigated.
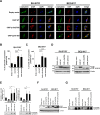
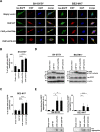
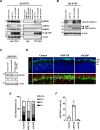
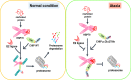
Results: We identified a novel heterozygous frameshift variant, c.832del (p.Glu278fs), in STUB1 in two patients from the same family. This rare mutation is located in the U-box of the carboxyl terminus of the Hsc70-interacting protein (CHIP) protein, which is encoded by STUB1. Further in vitro experiments demonstrated that this novel heterozygous STUB1 frameshift variant impairs the CHIP protein's activity and its interaction with the E2 ubiquitin ligase, UbE2D1, leading to neuronal accumulation of tau and α-synuclein, caspase-3 activation, and promoting cellular apoptosis through a dominant-negative pathogenic effect. The in vivo study revealed the influence of the CHIP expression level on the differentiation and migration of cerebellar granule neuron progenitors during cerebellar development.
Conclusions: Our findings provide clinical, genetic, and a mechanistic insight linking the novel heterozygous STUB1 frameshift mutation at the highly conserved U-box domain of CHIP as the cause of autosomal dominant SCA48. Our results further stress the importance of CHIP activity in neuronal protein homeostasis and cerebellar functions.
Keywords: Ataxia; CHIP; STUB1; Spinocerebellar ataxia type 48; Tau; α-Synuclein.
© 2021. The Author(s).
Conflict of interest statement
All authors report no competing interests.
Figures
References
-
- Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–4545. doi: 10.1128/MCB.19.6.4535. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

