Influence of B. subtilis 3NA mutations in spo0A and abrB on surfactin production in B. subtilis 168
- PMID: 34565366
- PMCID: PMC8474915
- DOI: 10.1186/s12934-021-01679-z
Influence of B. subtilis 3NA mutations in spo0A and abrB on surfactin production in B. subtilis 168
Abstract
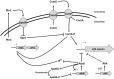
Background: Bacillus subtilis is a well-established host for a variety of bioproduction processes, with much interest focused on the production of biosurfactants such as the cyclic lipopeptide surfactin. Surfactin production is tightly intertwined with quorum sensing and regulatory cell differentiation processes. As previous studies have shown, a non-sporulating B. subtilis strain 3NA encoding a functional sfp locus but mutations in the spo0A and abrB loci, called JABs32, exhibits noticeably increased surfactin production capabilities. In this work, the impacts of introducing JABs32 mutations in the genes spo0A, abrB and abh from 3NA into strain KM1016, a surfactin-forming derivative of B. subtilis 168, was investigated. This study aims to show these mutations are responsible for the surfactin producing performance of strain JABs32 in fed-batch bioreactor cultivations.
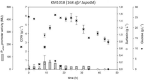
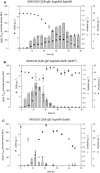
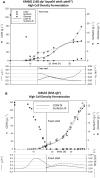
Results: Single and double mutant strains of B. subtilis KM1016 were constructed encoding gene deletions of spo0A, abrB and homologous abh. Furthermore, an elongated abrB version, called abrB*, as described for JABs32 was integrated. Single and combinatory mutant strains were analysed in respect of growth behaviour, native PsrfA promoter expression and surfactin production. Deletion of spo0A led to increased growth rates with lowered surfactin titers, while deletion or elongation of abrB resulted in lowered growth rates and high surfactin yields, compared to KM1016. The double mutant strains B. subtilis KM1036 and KM1020 encoding Δspo0A abrB* and Δspo0A ΔabrB were compared to reference strain JABs32, with KM1036 exhibiting similar production parameters and impeded cell growth and surfactin production for KM1020. Bioreactor fed-batch cultivations comparing a Δspo0A abrB* mutant of KM1016, KM681, with JABs32 showed a decrease of 32% in surfactin concentration.
Conclusions: The genetic differences of B. subtilis KM1016 and JABs32 give rise to new and improved fermentation methods through high cell density processes. Deletion of the spo0A locus was shown to be the reason for higher biomass concentrations. Only in combination with an elongation of abrB was this strain able to reach high surfactin titers of 18.27 g L-1 in fed-batch cultivations. This work shows, that a B. subtilis strain can be turned into a high cell density surfactin production strain by introduction of two mutations.
Keywords: AbrB; Bacillus subtilis; High cell density; Lipopeptide; Spo0A; Strain engineering; Surfactin.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N, Ishikawa S. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011;39:414–428. doi: 10.1093/nar/gkq780. - DOI - PMC - PubMed
-
- Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol Microbiol. 2005;57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

