Accelerated Barrier Repair in Human Skin Explants Induced with a Plant-Derived PPAR-α Activating Complex via Cooperative Interactions
- PMID: 34566418
- PMCID: PMC8458040
- DOI: 10.2147/CCID.S325967
Accelerated Barrier Repair in Human Skin Explants Induced with a Plant-Derived PPAR-α Activating Complex via Cooperative Interactions
Abstract
Background: Peroxisome proliferator-activated receptors (PPARs) govern epidermal lipid synthesis and metabolism. In skin, PPAR activation has been shown to regulate genes responsible for permeability barrier homeostasis, epidermal differentiation, lipid biosynthesis, and inflammation.
Objective: Given the known dermatologic benefits of PPARs, we set out to discover a naturally derived, multi-molecule complex that would be superior to the more commonly formulated conjugated linoleic acids (CLAs). We hypothesized that a complex may be capable of modulating PPAR-α by cooperative or multi-ligand binding interactions to accelerate skin barrier repair.
Methods: To achieve this, we assembled a novel PPAR-α agonist complex, referred to as RFV3, from a combination of small molecules routinely used in Ayurvedic medicine and accepted in cosmetic and topical over-the-counter dermatologic products. We tested RFV3's potential as a PPAR-α agonist by evaluating its transcriptional response, ligand binding affinity to PPAR-α, gene expression profiles and barrier repair properties in human skin explant models.
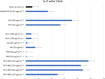
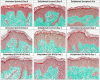
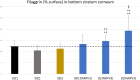
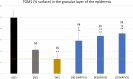
Results: We assembled RFV3 by solubilizing two standardized plant extracts in a suitable solvent and induced a significant transcriptional response in PPAR-α luciferase reporter assay. Furthermore, transcriptome profiling of RFV3-treated epidermal substitutes revealed expressed genes consistent with known targets of PPAR-α, including those involved in epidermal barrier repair. In addition, in silico modeling demonstrated differential co-binding affinities of RFV3 to PPAR-α compared with those of the endogenous ligands (CLAs) and a synthetic PPAR-α agonist. Lastly, delipidated skin explant models confirmed accelerated barrier repair activity with significant increases in ceramides, filaggrin and transglutaminase-1 after treatment.
Conclusion: These findings suggest that the RFV3 complex successfully mimics a PPAR-α agonist and induces synthesis of skin barrier lipids and proteins consistent with known PPAR pathways.
Keywords: PPAR-α; ceramides; cooperative binding; epidermal barrier; explants.
© 2021 Majewski et al.
Conflict of interest statement
Authors JC and TF are employees of Rodan & Fields, San Francisco, CA. Author GM was a consultant of Rodan & Fields during the research presented within and development of this manuscript. Mr GM and Dr JC report being inventors on patent application PPAR AGONIST COMPLEX AND METHODS OF USE, PCT/US2020/064798; Dr Timothy Falla discloses a patent PCT/US2020/064798 pending to Rodan + Fields. The authors report no other conflicts of interest in this work.
Figures






References
-
- Evans RM, Barish GD, Wang Y-X. PPARs and the complex journey to obesity. Nat Med. 2004;10(4):355–361. - PubMed
-
- Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–435. - PubMed
-
- Schmuth M, Moosbrugger-Martinz V, Blunder S, Dubrac S. Role of PPAR, LXR, and PXR in epidermal homeostasis and inflammation. Biochim Biophys Acta. 2014;1841(3):463–473. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

