Comprehensive analysis of abnormal expression, prognostic value and oncogenic role of the hub gene FN1 in pancreatic ductal adenocarcinoma via bioinformatic analysis and in vitro experiments
- PMID: 34567847
- PMCID: PMC8428264
- DOI: 10.7717/peerj.12141
Comprehensive analysis of abnormal expression, prognostic value and oncogenic role of the hub gene FN1 in pancreatic ductal adenocarcinoma via bioinformatic analysis and in vitro experiments
Abstract
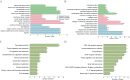
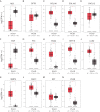
Background: Pancreatic ductal adenocarcinoma (PDAC) is one of the most commonly diagnosed cancers with a poor prognosis worldwide. Although the treatment of PDAC has made great progress in recent years, the therapeutic effects are still unsatisfactory. Methods. In this study, we identified differentially expressed genes (DEGs) between PDAC and normal pancreatic tissues based on four Gene Expression Omnibus (GEO) datasets (GSE15471, GSE16515, GSE28735 and GSE71729). A protein-protein interaction (PPI) network was established to evaluate the relationship between the DEGs and to screen hub genes. The expression levels of the hub genes were further validated through the Gene Expression Profiling Interactive Analysis (GEPIA), ONCOMINE and Human Protein Atlas (HPA) databases, as well as the validation GEO dataset GSE62452. Additionally, the prognostic values of the hub genes were evaluated by Kaplan-Meier plotter and the validation GEO dataset GSE62452. Finally, the mechanistic roles of the most remarkable hub genes in PDAC were examined through in vitro experiments.
Results: We identified the following nine hub genes by performing an integrated bioinformatics analysis: COL1A1, COL1A2, FN1, ITGA2, KRT19, LCN2, MMP9, MUC1 and VCAN. All of the hub genes were significantly upregulated in PDAC tissues compared with normal pancreatic tissues. Two hub genes (FN1 and ITGA2) were associated with poor overall survival (OS) rates in PDAC patients. Finally, in vitro experiments indicated that FN1 plays vital roles in PDAC cell proliferation, colony formation, apoptosis and the cell cycle.
Conclusions: In summary, we identified two hub genes that are associated with the expression and prognosis of PDAC. The oncogenic role of FN1 in PDAC was first illustrated by performing an integrated bioinformatic analysis and in vitro experiments. Our results provide a fundamental contribution for further research aimed finding novel therapeutic targets for overcoming PDAC.
Keywords: Bioinformatic analysis; FN1; I. vitro experiments; Pancreatic ductal adenocarcinoma.
© 2021 Lei et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Australian Pancreatic Cancer Genome I, Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

