Integrating 31-Gene Expression Profiling With Clinicopathologic Features to Optimize Cutaneous Melanoma Sentinel Lymph Node Metastasis Prediction
- PMID: 34568719
- PMCID: PMC8457832
- DOI: 10.1200/PO.21.00162
Integrating 31-Gene Expression Profiling With Clinicopathologic Features to Optimize Cutaneous Melanoma Sentinel Lymph Node Metastasis Prediction
Abstract
National guidelines recommend sentinel lymph node biopsy (SLNB) be offered to patients with > 10% likelihood of sentinel lymph node (SLN) positivity. On the other hand, guidelines do not recommend SLNB for patients with T1a tumors without high-risk features who have < 5% likelihood of a positive SLN. However, the decision to perform SLNB is less certain for patients with higher-risk T1 melanomas in which a positive node is expected 5%-10% of the time. We hypothesized that integrating clinicopathologic features with the 31-gene expression profile (31-GEP) score using advanced artificial intelligence techniques would provide more precise SLN risk prediction.
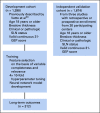
Methods: An integrated 31-GEP (i31-GEP) neural network algorithm incorporating clinicopathologic features with the continuous 31-GEP score was developed using a previously reported patient cohort (n = 1,398) and validated using an independent cohort (n = 1,674).
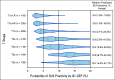
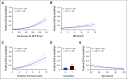
Results: Compared with other covariates in the i31-GEP, the continuous 31-GEP score had the largest likelihood ratio (G2 = 91.3, P < .001) for predicting SLN positivity. The i31-GEP demonstrated high concordance between predicted and observed SLN positivity rates (linear regression slope = 0.999). The i31-GEP increased the percentage of patients with T1-T4 tumors predicted to have < 5% SLN-positive likelihood from 8.5% to 27.7% with a negative predictive value of 98%. Importantly, for patients with T1 tumors originally classified with a likelihood of SLN positivity of 5%-10%, the i31-GEP reclassified 63% of cases as having < 5% or > 10% likelihood of positive SLN, for a more precise, personalized, and clinically actionable SLN-positive likelihood estimate.
Conclusion: These data suggest the i31-GEP could reduce the number of SLNBs performed by identifying patients with likelihood under the 5% threshold for performance of SLNB and improve the yield of positive SLNBs by identifying patients more likely to have a positive SLNB.
Trial registration: ClinicalTrials.gov NCT02355587 NCT02355574.
© 2021 by American Society of Clinical Oncology.
Conflict of interest statement
John T. Vetto Employment: Gilead Sciences (I) Stock and Other Ownership Interests: Gilead Sciences (I), Roche/Genentech (I) Speakers' Bureau: Castle Biosciences Travel, Accommodations, Expenses: Castle Biosciences No other potential conflicts of interest were reported. John T. Vetto Employment: Gilead Sciences (I) Stock and Other Ownership Interests: Gilead Sciences (I), Roche/Genentech (I) Speakers' Bureau: Castle Biosciences Travel, Accommodations, Expenses: Castle Biosciences No other potential conflicts of interest were reported.
Figures



References
-
- Swetter SM Thompson JA Coit DG, et al. : NCCN Clinical Practice Guidelines in Oncology. Cutaneous Melanoma. Version 3.2020. Plymouth Meeting, PA, National Comprehensive Cancer Network, 2020
-
- Friedman C Lyon M Torphy RJ, et al. : A nomogram to predict node positivity in patients with thin melanomas helps inform shared patient decision making. J Surg Oncol 120:1276-1283, 2019 - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical

