Chromosome-scale assembly of wild barley accession "OUH602"
- PMID: 34568912
- PMCID: PMC8473966
- DOI: 10.1093/g3journal/jkab244
Chromosome-scale assembly of wild barley accession "OUH602"
Abstract
Barley (Hordeum vulgare) was domesticated from its wild ancestral form ca. 10,000 years ago in the Fertile Crescent and is widely cultivated throughout the world, except for in tropical areas. The genome size of both cultivated barley and its conspecific wild ancestor is approximately 5 Gb. High-quality chromosome-level assemblies of 19 cultivated and one wild barley genotype were recently established by pan-genome analysis. Here, we release another equivalent short-read assembly of the wild barley accession "OUH602." A series of genetic and genomic resources were developed for this genotype in prior studies. Our assembly contains more than 4.4 Gb of sequence, with a scaffold N50 value of over 10 Mb. The haplotype shows high collinearity with the most recently updated barley reference genome, "Morex" V3, with some inversions. Gene projections based on "Morex" gene models revealed 46,807 protein-coding sequences and 43,375 protein-coding genes. Alignments to publicly available sequences of bacterial artificial chromosome (BAC) clones of "OUH602" confirm the high accuracy of the assembly. Since more loci of interest have been identified in "OUH602," the release of this assembly, with detailed genomic information, should accelerate gene identification and the utilization of this key wild barley accession.
Keywords: Hordeum vulgare ssp. spontaneum; OUH602; genome assembly; pseudomolecules; wild barley.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures
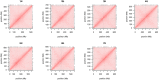
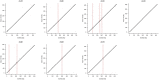
References
-
- Bailly-Bechet M, Haudry A, Lerat E.. 2014. “One code to find them all”: a Perl tool to conveniently parse RepeatMasker output files. Mobile DNA. 5:13.doi:10.1186/1759-8753-5-13.
-
- Bothmer R, von K, Sato T, Komatsuda S, Yasuda Fischbeck G.. 2003. The domestication of cultivated barley. In: von Bothmer R, van Hintum Knüpffer TH, Sato H K, editors, Diversity in Barley (Hordeum vulgare),. The Netherlands: Elsevier Science B. V, p. 9–27.
-
- Clavijo JB, Accinelli GG, Wright J, Heavens D, Barr K.. et al. 2017. W2RAP: a pipeline for high quality, robust assemblies of large complex genomes from short read data. bioRxiv. 10.1101/110999
-
- Dvorak J, McGuire PE, Cassidy B.. 1988. Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome. 30:680–689. doi:10.1139/g88-115.
-
- Gyenis L, Yun SJ, Smith KP, Steffenson BJ, Bossolini E, et al.2007. Genetic architecture of quantitative trait loci associated with morphological and agronomic trait differences in a wild by cultivated barley cross. Genome. 50:714–723. doi:10.1139/g07-054. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
