Identification of a dual acting SARS-CoV-2 proteases inhibitor through in silico design and step-by-step biological characterization
- PMID: 34571172
- PMCID: PMC8457654
- DOI: 10.1016/j.ejmech.2021.113863
Identification of a dual acting SARS-CoV-2 proteases inhibitor through in silico design and step-by-step biological characterization
Abstract
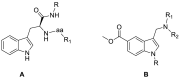
COVID-19 pandemic, starting from the latest 2019, and caused by SARS-CoV-2 pathogen, led to the hardest health-socio-economic disaster in the last century. Despite the tremendous scientific efforts, mainly focused on the development of vaccines, identification of potent and efficient anti-SARS-CoV-2 therapeutics still represents an unmet need. Remdesivir, an anti-Ebola drug selected from a repurposing campaign, is the only drug approved, so far, for the treatment of the infection. Nevertheless, WHO in later 2020 has recommended against its use in COVID-19. In the present paper, we describe a step-by-step in silico design of a small library of compounds as main protease (Mpro) inhibitors. All the molecules were screened by an enzymatic assay on Mpro and, then, cellular activity was evaluated using Vero cells viral infection model. The cellular screening disclosed compounds 29 and 34 as in-vitro SARS-CoV-2 replication inhibitors at non-toxic concentrations (0.32 < EC50 < 5.98 μM). To rationalize these results, additional in-vitro assays were performed, focusing on papain like protease (PLpro) and spike protein (SP) as potential targets for the synthesized molecules. This pharmacological workflow allowed the identification of compound 29, as a dual acting SARS-CoV-2 proteases inhibitor featuring micromolar inhibitory potency versus Mpro (IC50 = 1.72 μM) and submicromolar potency versus PLpro (IC50 = 0.67 μM), and of compound 34 as a selective SP inhibitor (IC50 = 3.26 μM).
Keywords: Biophysical assays; Cellular characterization; Enzymatic assays; In-silico design; SARS-CoV-2 proteases dual inhibitor.
Copyright © 2021 Elsevier Masson SAS. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease.J Med Chem. 2022 Jun 9;65(11):7561-7580. doi: 10.1021/acs.jmedchem.2c00303. Epub 2022 May 27. J Med Chem. 2022. PMID: 35620927 Free PMC article. Review.
-
Peptidomimetics as potent dual SARS-CoV-2 cathepsin-L and main protease inhibitors: In silico design, synthesis and pharmacological characterization.Eur J Med Chem. 2024 Feb 15;266:116128. doi: 10.1016/j.ejmech.2024.116128. Epub 2024 Jan 9. Eur J Med Chem. 2024. PMID: 38232463
-
Kinetic Characterization and Inhibitor Screening for the Proteases Leading to Identification of Drugs against SARS-CoV-2.Antimicrob Agents Chemother. 2021 Mar 18;65(4):e02577-20. doi: 10.1128/AAC.02577-20. Print 2021 Mar 18. Antimicrob Agents Chemother. 2021. PMID: 33526482 Free PMC article.
-
Non-Toxic Dimeric Peptides Derived from the Bothropstoxin-I Are Potent SARS-CoV-2 and Papain-like Protease Inhibitors.Molecules. 2021 Aug 12;26(16):4896. doi: 10.3390/molecules26164896. Molecules. 2021. PMID: 34443484 Free PMC article.
-
Different Aspects of Emetine's Capabilities as a Highly Potent SARS-CoV-2 Inhibitor against COVID-19.ACS Pharmacol Transl Sci. 2022 May 23;5(6):387-399. doi: 10.1021/acsptsci.2c00045. eCollection 2022 Jun 10. ACS Pharmacol Transl Sci. 2022. PMID: 35702393 Free PMC article. Review.
Cited by
-
SARS-COV-2 Coronavirus Papain-like Protease PLpro as an Antiviral Target for Inhibitors of Active Site and Protein-Protein Interactions.Biophysics (Oxf). 2022;67(6):902-912. doi: 10.1134/S0006350922060082. Epub 2023 Mar 3. Biophysics (Oxf). 2022. PMID: 36883182 Free PMC article.
-
Discovery of PLpro and Mpro Inhibitors for SARS-CoV-2.ACS Omega. 2023 Jun 14;8(25):22603-22612. doi: 10.1021/acsomega.3c01110. eCollection 2023 Jun 27. ACS Omega. 2023. PMID: 37387790 Free PMC article.
-
Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease.J Med Chem. 2022 Jun 9;65(11):7561-7580. doi: 10.1021/acs.jmedchem.2c00303. Epub 2022 May 27. J Med Chem. 2022. PMID: 35620927 Free PMC article. Review.
-
Main and papain-like proteases as prospective targets for pharmacological treatment of coronavirus SARS-CoV-2.RSC Adv. 2023 Dec 6;13(50):35500-35524. doi: 10.1039/d3ra06479d. eCollection 2023 Nov 30. RSC Adv. 2023. PMID: 38077980 Free PMC article. Review.
-
The SARS-CoV-2 main protease (Mpro): Structure, function, and emerging therapies for COVID-19.MedComm (2020). 2022 Jul 14;3(3):e151. doi: 10.1002/mco2.151. eCollection 2022 Sep. MedComm (2020). 2022. PMID: 35845352 Free PMC article. Review.
References
-
- Dhama K., Patel S.K., Sharun K., Pathak M., Tiwari R., Yatoo M.I., Malik Y.S., Sah R., Rabaan A.A., Panwar P.K., Singh K.P., Michalak I., Chaicumpa W., Martinez-Pulgarin D.F., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2 jumping the species barrier: zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Trav. Med. Infect. Dis. 2020;37:101830. - PMC - PubMed
-
- Triggle C.R., Bansal D., Ding H., Islam M.M., Farag E., Hadi H.A., Sultan A.A. A comprehensive review of viral characteristics, transmission, pathophysiology, immune response, and management of SARS-CoV-2 and COVID-19 as a basis for controlling the pandemic. Front. Immunol. 2021;12:631139. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Chemical Information
Miscellaneous

