A Universal Approach to Analyzing Transmission Electron Microscopy with ImageJ
- PMID: 34571826
- PMCID: PMC8465115
- DOI: 10.3390/cells10092177
A Universal Approach to Analyzing Transmission Electron Microscopy with ImageJ
Abstract
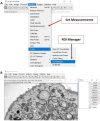
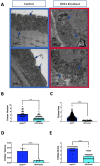
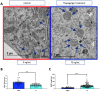
Transmission electron microscopy (TEM) is widely used as an imaging modality to provide high-resolution details of subcellular components within cells and tissues. Mitochondria and endoplasmic reticulum (ER) are organelles of particular interest to those investigating metabolic disorders. A straightforward method for quantifying and characterizing particular aspects of these organelles would be a useful tool. In this protocol, we outline how to accurately assess the morphology of these important subcellular structures using open source software ImageJ, originally developed by the National Institutes of Health (NIH). Specifically, we detail how to obtain mitochondrial length, width, area, and circularity, in addition to assessing cristae morphology and measuring mito/endoplasmic reticulum (ER) interactions. These procedures provide useful tools for quantifying and characterizing key features of sub-cellular morphology, leading to accurate and reproducible measurements and visualizations of mitochondria and ER.
Keywords: ImageJ; Mitochondria Endoplasmic Reticulum Contacts (MERCs); TEM analysis; TEM quantification; cristae; image analysis; image processing; mitochondria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

