Autophagy-Associated IL-15 Production Is Involved in the Pathogenesis of Leprosy Type 1 Reaction
- PMID: 34571865
- PMCID: PMC8468917
- DOI: 10.3390/cells10092215
Autophagy-Associated IL-15 Production Is Involved in the Pathogenesis of Leprosy Type 1 Reaction
Abstract
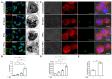
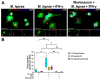
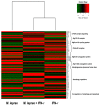
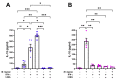
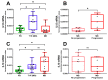
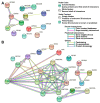
Leprosy reactional episodes are acute inflammatory events that may occur during the clinical course of the disease. Type 1 reaction (T1R) is associated with an increase in neural damage, and the understanding of the molecular pathways related to T1R onset is pivotal for the development of strategies that may effectively control the reaction. Interferon-gamma (IFN-γ) is a key cytokine associated with T1R onset and is also associated with autophagy induction. Here, we evaluated the modulation of the autophagy pathway in Mycobacterium leprae-stimulated cells in the presence or absence of IFN-γ. We observed that IFN-γ treatment promoted autophagy activation and increased the expression of genes related to the formation of phagosomes, autophagy regulation and function, or lysosomal pathways in M. leprae-stimulated cells. IFN-γ increased interleukin (IL)-15 secretion in M. leprae-stimulated THP-1 cells in a process associated with autophagy activation. We also observed higher IL15 gene expression in multibacillary (MB) patients who later developed T1R during clinical follow-up when compared to MB patients who did not develop the episode. By overlapping gene expression patterns, we observed 13 common elements shared between T1R skin lesion cells and THP-1 cells stimulated with both M. leprae and IFN-γ. Among these genes, the autophagy regulator Translocated Promoter Region, Nuclear Basket Protein (TPR) was significantly increased in T1R cells when compared with non-reactional MB cells. Overall, our results indicate that IFN-γ may induce a TPR-mediated autophagy transcriptional program in M. leprae-stimulated cells similar to that observed in skin cells during T1R by a pathway that involves IL-15 production, suggesting the involvement of this cytokine in the pathogenesis of T1R.
Keywords: IL-15; Mycobacterium leprae; autophagy; leprosy or T1R; lysosomes; macrophage or THP-1; phagosome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Lockwood D.N.J., Lucas S.B., Desikan K.V., Ebenezer G., Suneetha S., Nicholls P. The histological diagnosis of leprosy type 1 reactions: Identification of key variables and an analysis of the process of histological diagnosis. J. Clin. Pathol. 2008;61:595–600. doi: 10.1136/jcp.2007.053389. - DOI - PubMed
-
- Jih M.H., Kimyai-Asadi A., Levis W.R. Reversal reaction to Hansen’s disease. J. Drugs Dermatol. 2002;1:70–71. - PubMed
-
- Smith W.C., Anderson A.M., Withington S.G., van Brakel W.H., Croft R.P., Nicholls P.G., Richardus J.H. Steroid prophylaxis for prevention of nerve function impairment in leprosy: Randomised placebo controlled trial (TRIPOD 1) BMJ. 2004;328:1459. doi: 10.1136/bmj.38107.645926.AE. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

