Mutations of SARS-CoV-2 RBD May Alter Its Molecular Structure to Improve Its Infection Efficiency
- PMID: 34572486
- PMCID: PMC8466379
- DOI: 10.3390/biom11091273
Mutations of SARS-CoV-2 RBD May Alter Its Molecular Structure to Improve Its Infection Efficiency
Abstract
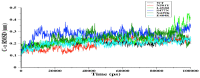
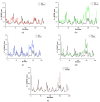
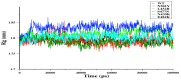
The receptor-binding domain (RBD) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mediates the viral-host interaction and is a target for most neutralizing antibodies. Nevertheless, SARS-CoV-2 RBD mutations pose a threat due to their role in host cell entry via the human angiotensin-converting enzyme 2 receptor that might strengthen SARS-CoV-2 infectivity, viral load, or resistance against neutralizing antibodies. To understand the molecular structural link between RBD mutations and infectivity, the top five mutant RBDs (i.e., N501Y, E484K L452R, S477N, and N439K) were selected based on their recorded case numbers. These mutants along with wild-type (WT) RBD were studied through all-atom molecular dynamics (MD) simulations of 100 ns. The principal component analysis and the free energy landscape were used too. Interestingly, N501Y, N439K, and E484K mutations were observed to increase the rigidity in some RBD regions while increasing the flexibility of the receptor-binding motif (RBM) region, suggesting a compensation of the entropy penalty. However, S477N and L452R RBDs were observed to increase the flexibility of the RBM region while maintaining similar flexibility in other RBD regions in comparison to WT RBD. Therefore, both mutations (especially S477N) might destabilize the RBD structure, as loose conformation compactness was observed. The destabilizing effect of S477N RBD was consistent with previous work on S477N mutation. Finally, the free energy landscape results showed that mutations changed WT RBD conformation while local minima were maintained for all mutant RBDs. In conclusion, RBD mutations definitely impact the WT RBD structure and conformation as well as increase the binding affinity to angiotensin-converting enzyme receptor.
Keywords: RBD flexibility; SARS-CoV-2; free energy landscape; molecular dynamics simulations; mutant RBDs; principle component analysis; wild-type RBD.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- WHO Coronavirus (COVID-19) Dashboard. [(accessed on 6 June 2021)]. Available online: https://covid19.who.int.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

