Human-Associated Methicillin-Resistant Staphylococcus aureus Clonal Complex 80 Isolated from Cattle and Aquatic Environments
- PMID: 34572619
- PMCID: PMC8468323
- DOI: 10.3390/antibiotics10091038
Human-Associated Methicillin-Resistant Staphylococcus aureus Clonal Complex 80 Isolated from Cattle and Aquatic Environments
Abstract
Background: Human-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) has mainly been reported in South African pig and chicken farms. The prevalence of antibiotic-resistant genes (ARGs), virulence factors (VFs), and multilocus sequence types (MLSTs) associated with HA-MRSA in cattle farms has not been reported. Consequently, this study characterised LA-MRSA and its spread from cattle farms into the environment.
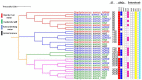
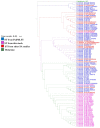
Method: Husbandry soil (HS), nearby river water (NRW), animal manure (AM) and animal drinking water (ADW) were collected on and around a cattle farm. Presumptive MRSA isolates were identified from these samples using CHROMagar media and genotyped as MRSA sequence types (STs), selected ARGs, and VFs, using polymerase chain reaction. An MLST-based dendrogram was generated to link the farm MRSA strains with those in a nearby river.
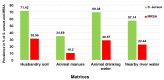
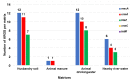
Results: The prevalence of MRSA was 30.61% for HS, 28.57% for ADW, 22.44% for NRW, and 10.20% for AM. Isolates from HS harboured the highest number of resistant genes, with 100% for mecA, 91.66% for ermA, and 58.33% for blaZ. However, no ermC or tetM genes were detected. MRSA isolates from AM harboured the lowest number of resistant genes. Only sec and seq enterotoxins were found in all the assessed MRSA isolates. MRSA from the farm revealed six STs (ST80, ST728, ST1931, ST2030, ST3247, and ST5440); all of STs belonged to clonal complex 80 (CC80). An MLST-based dendrogram based on the concatenated sequences of MLST genes under the maximum likelihood criterion revealed four clades of amalgamated MRSA isolates from various livestock environmental matrices, including the NRW.
Conclusion: The results suggest that livestock environmental matrices might be reservoirs of MRSA that could subsequently disseminate through runoff to pollute water resources. Therefore, continued surveillance of HA-MRSA in livestock environments is warranted.
Keywords: husbandry soil; livestock; manure; methicillin-resistant Staphylococcus aureus; water.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Prevalence and Molecular Characterisation of Extended-Spectrum Beta-Lactamase-Producing Shiga Toxin-Producing Escherichia coli, from Cattle Farm to Aquatic Environments.Pathogens. 2022 Jun 10;11(6):674. doi: 10.3390/pathogens11060674. Pathogens. 2022. PMID: 35745529 Free PMC article.
-
Bovine mastitis Staphylococcus aureus: antibiotic susceptibility profile, resistance genes and molecular typing of methicillin-resistant and methicillin-sensitive strains in China.Infect Genet Evol. 2015 Apr;31:9-16. doi: 10.1016/j.meegid.2014.12.039. Epub 2015 Jan 9. Infect Genet Evol. 2015. PMID: 25582604
-
Unexpected sequence types in livestock associated methicillin-resistant Staphylococcus aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China.Vet Microbiol. 2009 Nov 18;139(3-4):405-9. doi: 10.1016/j.vetmic.2009.06.014. Epub 2009 Jun 21. Vet Microbiol. 2009. PMID: 19608357
-
Methicillin-resistant Staphylococcus aureus: a controversial food-borne pathogen.Lett Appl Microbiol. 2017 Jun;64(6):409-418. doi: 10.1111/lam.12735. Epub 2017 May 3. Lett Appl Microbiol. 2017. PMID: 28304109 Review.
-
Methicillin resistance in Staphylococcus isolates: the "mec alphabet" with specific consideration of mecC, a mec homolog associated with zoonotic S. aureus lineages.Int J Med Microbiol. 2014 Oct;304(7):794-804. doi: 10.1016/j.ijmm.2014.06.007. Epub 2014 Jun 28. Int J Med Microbiol. 2014. PMID: 25034857 Review.
Cited by
-
Methicillin-resistant Staphylococcus aureus prevalence in food-producing animals and food products in Saudi Arabia: A review.Vet World. 2024 Aug;17(8):1753-1764. doi: 10.14202/vetworld.2024.1753-1764. Epub 2024 Aug 13. Vet World. 2024. PMID: 39328450 Free PMC article. Review.
-
A 6-Year Update on the Diversity of Methicillin-Resistant Staphylococcus aureus Clones in Africa: A Systematic Review.Front Microbiol. 2022 May 3;13:860436. doi: 10.3389/fmicb.2022.860436. eCollection 2022. Front Microbiol. 2022. PMID: 35591993 Free PMC article. Review.
-
Prevalence and Molecular Characterisation of Extended-Spectrum Beta-Lactamase-Producing Shiga Toxin-Producing Escherichia coli, from Cattle Farm to Aquatic Environments.Pathogens. 2022 Jun 10;11(6):674. doi: 10.3390/pathogens11060674. Pathogens. 2022. PMID: 35745529 Free PMC article.
-
Identification, Comparison, and Profiling of Selected Diarrhoeagenic Pathogens from Diverse Water Sources and Human and Animal Faeces Using Whole-Genome Sequencing.Microorganisms. 2025 Jun 12;13(6):1373. doi: 10.3390/microorganisms13061373. Microorganisms. 2025. PMID: 40572261 Free PMC article.
References
-
- Amarasiri M., Sano D., Suzuki S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2019:1–44. doi: 10.1080/10643389.2019.1692611. - DOI
-
- Edokpayi J.N., Rogawski E.T., Kahler D.M., Hill C.L., Reynolds C., Nyathi E., Smith J.A., Odiyo J.O., Samie A., Bessong P., et al. Challenges to sustainable safe drinking water: A case study of water quality and use across seasons in rural communities in Limpopo Province, South Africa. Water. 2018;10:159. doi: 10.3390/w10020159. - DOI - PMC - PubMed
-
- Verlicchi P., Grillini V. Surface Water and Groundwater Quality in South Africa and Mozambique—Analysis of the Most Critical Pollutants for Drinking Purposes and Challenges in Water Treatment Selection. Water. 2020;12:305. doi: 10.3390/w12010305. - DOI
-
- Van den Honert M.S., Gouws P.A., Hoffman L.C. Importance and implications of antibiotic resistance development in livestock and wildlife farming in South Africa: A Review. South Afr. J. Anim. Sci. 2018;48:401–412. doi: 10.4314/sajas.v48i3.1. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

