Saccharomyces Cerevisiae Var Boulardii CNCM I-1079 Reduces Expression of Genes Involved in Inflammatory Response in Porcine Cells Challenged by Enterotoxigenic E. Coli and Influences Bacterial Communities in an In Vitro Model of the Weaning Piglet Colon
- PMID: 34572682
- PMCID: PMC8467900
- DOI: 10.3390/antibiotics10091101
Saccharomyces Cerevisiae Var Boulardii CNCM I-1079 Reduces Expression of Genes Involved in Inflammatory Response in Porcine Cells Challenged by Enterotoxigenic E. Coli and Influences Bacterial Communities in an In Vitro Model of the Weaning Piglet Colon
Abstract
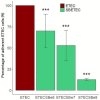
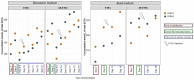
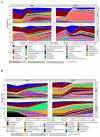
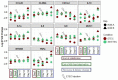
Enterotoxigenic Escherichia coli (ETEC) is the main infectious agent responsible for piglet post-weaning diarrhea with high mortality rates. Antimicrobials represent the current principal strategy for treating ETEC infections in pig farms, but the occurrence of multi-resistant bacterial strains has considerably increased in the last decades. Thus, finding non-antibiotic alternatives becomes a real emergency. In this context, we investigated the effect of a live yeast strain, Saccharomyces cerevisiae var boulardii CNCM I-1079 (SB) in an in vitro model of the weaning piglet colon implemented with a mucus phase (MPigut-IVM) inoculated with ETEC and coupled with an intestinal porcine cell line IPI-2I. We showed that SB was able to modulate the in vitro microbiota through an increase in Bacteroidiaceae and a decrease in Prevotellaceae families. Effluents collected from the SB treated bioreactors were able to mitigate the expression level of genes encoding non-gel forming mucins, tight junction proteins, innate immune pathway, and pro-inflammatory response in IPI-2I cells. Furthermore, SB exerted a significant protective effect against ETEC adhesion on porcine IPEC-J2 intestinal cells in a dose-dependent manner and showed a positive effect on ETEC-challenged IPEC-J2 by lowering expression of genes involved in pro-inflammatory immune responses. Our results showed that the strain SB CNCM I-1079 could prevent microbiota dysbiosis associated with weaning and protect porcine enterocytes from ETEC infections by reducing bacterial adhesion and modulating the inflammatory response.
Keywords: ETEC; in vitro model of colonic microbiota; intestinal cells; piglet; probiotics; weaning.
Conflict of interest statement
F.C.D. and R.G. are employees of Lallemand SAS. The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Pathogen Challenge and Dietary Shift Alter Microbiota Composition and Activity in a Mucin-Associated in vitro Model of the Piglet Colon (MPigut-IVM) Simulating Weaning Transition.Front Microbiol. 2021 Jul 19;12:703421. doi: 10.3389/fmicb.2021.703421. eCollection 2021. Front Microbiol. 2021. PMID: 34349744 Free PMC article.
-
Effect of Saccharomyces cerevisiae var. Boulardii and β-galactomannan oligosaccharide on porcine intestinal epithelial and dendritic cells challenged in vitro with Escherichia coli F4 (K88).Vet Res. 2012 Jan 25;43(1):4. doi: 10.1186/1297-9716-43-4. Vet Res. 2012. PMID: 22277078 Free PMC article.
-
Anti-infectious properties of the probiotic Saccharomyces cerevisiae CNCM I-3856 on enterotoxigenic E. coli (ETEC) strain H10407.Appl Microbiol Biotechnol. 2018 Jul;102(14):6175-6189. doi: 10.1007/s00253-018-9053-y. Epub 2018 May 25. Appl Microbiol Biotechnol. 2018. PMID: 29802478
-
Diversity of Saccharomyces boulardii CNCM I-745 mechanisms of action against intestinal infections.World J Gastroenterol. 2019 May 14;25(18):2188-2203. doi: 10.3748/wjg.v25.i18.2188. World J Gastroenterol. 2019. PMID: 31143070 Free PMC article. Review.
-
Saccharomyces boulardii CNCM I-745 supports regeneration of the intestinal microbiota after diarrheic dysbiosis - a review.Clin Exp Gastroenterol. 2015 Aug 14;8:237-55. doi: 10.2147/CEG.S85574. eCollection 2015. Clin Exp Gastroenterol. 2015. PMID: 26316791 Free PMC article. Review.
Cited by
-
Intestinal fungi biogeography, succession and its association with diarrhea in pigs.J Anim Sci Biotechnol. 2025 Jun 4;16(1):80. doi: 10.1186/s40104-025-01206-9. J Anim Sci Biotechnol. 2025. PMID: 40462211 Free PMC article.
-
Emerging relevance of cell wall components from non-conventional yeasts as functional ingredients for the food and feed industry.Curr Res Food Sci. 2023 Sep 30;7:100603. doi: 10.1016/j.crfs.2023.100603. eCollection 2023. Curr Res Food Sci. 2023. PMID: 37840697 Free PMC article. Review.
-
Cultivating complexity: Advancements in establishing in vitro models for the mucus-adhering gut microbiota.Microb Biotechnol. 2024 Oct;17(10):e70036. doi: 10.1111/1751-7915.70036. Microb Biotechnol. 2024. PMID: 39435730 Free PMC article. Review.
-
Probiotic Saccharomyces boulardii Against Cronobacter sakazakii Infection: In Vitro and In Vivo Studies.Probiotics Antimicrob Proteins. 2025 Mar 21. doi: 10.1007/s12602-025-10524-3. Online ahead of print. Probiotics Antimicrob Proteins. 2025. PMID: 40113720
References
LinkOut - more resources
Full Text Sources

