Precision Oncology with Drugs Targeting the Replication Stress, ATR, and Schlafen 11
- PMID: 34572827
- PMCID: PMC8465591
- DOI: 10.3390/cancers13184601
Precision Oncology with Drugs Targeting the Replication Stress, ATR, and Schlafen 11
Abstract
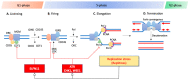
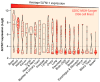
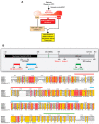
Precision medicine aims to implement strategies based on the molecular features of tumors and optimized drug delivery to improve cancer diagnosis and treatment. DNA replication is a logical approach because it can be targeted by a broad range of anticancer drugs that are both clinically approved and in development. These drugs increase deleterious replication stress (RepStress); however, how to selectively target and identify the tumors with specific molecular characteristics are unmet clinical needs. Here, we provide background information on the molecular processes of DNA replication and its checkpoints, and discuss how to target replication, checkpoint, and repair pathways with ATR inhibitors and exploit Schlafen 11 (SLFN11) as a predictive biomarker.
Keywords: ATR; Adavoserib; Camptothecin; Ceralasertib; PARP; Rucaparib; berzosertib; cisplatin; niraparib; olaparib; replication stress; schlafen 11; talazoparib; temozolomide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Kohn K.W. Beyond DNA cross-linking: History and prospects of DNA-targeted cancer treatment—Fifteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1996;56:5533–5546. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

