Enhanced Directed Random Walk for the Identification of Breast Cancer Prognostic Markers from Multiclass Expression Data
- PMID: 34573857
- PMCID: PMC8472068
- DOI: 10.3390/e23091232
Enhanced Directed Random Walk for the Identification of Breast Cancer Prognostic Markers from Multiclass Expression Data
Abstract
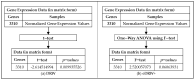
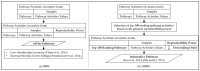
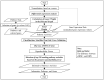
Artificial intelligence in healthcare can potentially identify the probability of contracting a particular disease more accurately. There are five common molecular subtypes of breast cancer: luminal A, luminal B, basal, ERBB2, and normal-like. Previous investigations showed that pathway-based microarray analysis could help in the identification of prognostic markers from gene expressions. For example, directed random walk (DRW) can infer a greater reproducibility power of the pathway activity between two classes of samples with a higher classification accuracy. However, most of the existing methods (including DRW) ignored the characteristics of different cancer subtypes and considered all of the pathways to contribute equally to the analysis. Therefore, an enhanced DRW (eDRW+) is proposed to identify breast cancer prognostic markers from multiclass expression data. An improved weight strategy using one-way ANOVA (F-test) and pathway selection based on the greatest reproducibility power is proposed in eDRW+. The experimental results show that the eDRW+ exceeds other methods in terms of AUC. Besides this, the eDRW+ identifies 294 gene markers and 45 pathway markers from the breast cancer datasets with better AUC. Therefore, the prognostic markers (pathway markers and gene markers) can identify drug targets and look for cancer subtypes with clinically distinct outcomes.
Keywords: ANOVA; breast cancer; directed random walk; microarray analysis; multiclass; pathway selection; prognostic markers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
An Entropy-Based Directed Random Walk for Cancer Classification Using Gene Expression Data Based on Bi-Random Walk on Two Separated Networks.Genes (Basel). 2023 Feb 24;14(3):574. doi: 10.3390/genes14030574. Genes (Basel). 2023. PMID: 36980844 Free PMC article.
-
Topologically inferring risk-active pathways toward precise cancer classification by directed random walk.Bioinformatics. 2013 Sep 1;29(17):2169-77. doi: 10.1093/bioinformatics/btt373. Epub 2013 Jul 10. Bioinformatics. 2013. PMID: 23842813
-
Mixture classification model based on clinical markers for breast cancer prognosis.Artif Intell Med. 2010 Feb-Mar;48(2-3):129-37. doi: 10.1016/j.artmed.2009.07.008. Epub 2009 Dec 14. Artif Intell Med. 2010. PMID: 20005686
-
Triple-negative breast cancer: current state of the art.Tumori. 2010 Nov-Dec;96(6):875-88. Tumori. 2010. PMID: 21388048 Review.
-
[Breast cancer: new concepts in classification].Rev Bras Ginecol Obstet. 2008 Jan;30(1):42-7. doi: 10.1590/s0100-72032008000100008. Rev Bras Ginecol Obstet. 2008. PMID: 19142542 Review. Portuguese.
Cited by
-
Identifying driver modules based on multi-omics biological networks in prostate cancer.IET Syst Biol. 2022 Dec;16(6):187-200. doi: 10.1049/syb2.12050. Epub 2022 Aug 30. IET Syst Biol. 2022. PMID: 36039671 Free PMC article.
References
-
- Mohapatra P., Chakravarty S., Dash P. Microarray medical data classification using kernel ridge regression and modified cat swarm optimization based gene selection system. Swarm Evol. Comput. 2016;28:144–160. doi: 10.1016/j.swevo.2016.02.002. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

