RT-LAMP CRISPR-Cas12/13-Based SARS-CoV-2 Detection Methods
- PMID: 34573987
- PMCID: PMC8467512
- DOI: 10.3390/diagnostics11091646
RT-LAMP CRISPR-Cas12/13-Based SARS-CoV-2 Detection Methods
Abstract
Coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has attracted public attention. The gold standard for diagnosing COVID-19 is reverse transcription-quantitative polymerase chain reaction (RT-qPCR). However, RT-qPCR can only be performed in centralized laboratories due to the requirement for advanced laboratory equipment and qualified workers. In the last decade, clustered regularly interspaced short palindromic repeats (CRISPR) technology has shown considerable promise in the development of rapid, highly sensitive, and specific molecular diagnostic methods that do not require complicated instrumentation. During the current COVID-19 pandemic, there has been growing interest in using CRISPR-based diagnostic techniques to develop rapid and accurate assays for detecting SARS-CoV-2. In this work, we review and summarize reverse-transcription loop-mediated isothermal amplification (RT-LAMP) CRISPR-based diagnostic techniques for detecting SARS-CoV-2.
Keywords: COVID-19; CRISPR; Cas12; Cas13; RT-LAMP; SARS-CoV-2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
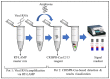
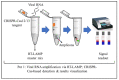


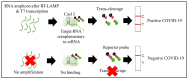
References
-
- Phua J., Weng L., Ling L., Egi M., Lim C.-M., Divatia J.V., Shrestha B.R., Arabi Y.M., Ng J., Gomersall C.D., et al. Intensive care management of coronavirus disease 2019 (COVID-19): Challenges and recommendations. Lancet Respir. Med. 2020;8:506–517. doi: 10.1016/S2213-2600(20)30161-2. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

