Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods
- PMID: 34575619
- PMCID: PMC8472571
- DOI: 10.3390/jpm11090842
Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods
Abstract
Radiomics converts medical images into mineable data via a high-throughput extraction of quantitative features used for clinical decision support. However, these radiomic features are susceptible to variation across scanners, acquisition protocols, and reconstruction settings. Various investigations have assessed the reproducibility and validation of radiomic features across these discrepancies. In this narrative review, we combine systematic keyword searches with prior domain knowledge to discuss various harmonization solutions to make the radiomic features more reproducible across various scanners and protocol settings. Different harmonization solutions are discussed and divided into two main categories: image domain and feature domain. The image domain category comprises methods such as the standardization of image acquisition, post-processing of raw sensor-level image data, data augmentation techniques, and style transfer. The feature domain category consists of methods such as the identification of reproducible features and normalization techniques such as statistical normalization, intensity harmonization, ComBat and its derivatives, and normalization using deep learning. We also reflect upon the importance of deep learning solutions for addressing variability across multi-centric radiomic studies especially using generative adversarial networks (GANs), neural style transfer (NST) techniques, or a combination of both. We cover a broader range of methods especially GANs and NST methods in more detail than previous reviews.
Keywords: deep learning; feature reproducibility; harmonization; medical imaging; radiomics.
Conflict of interest statement
Dutch patent filed by A.C. titled ‘Method of processing medical images by an analysis system for enabling radiomics signature analysis’ Patent no. P127348NL00.
Figures

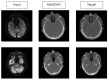
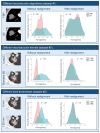

References
-
- Aerts H.J.W.L., Velazquez E.R., Leijenaar R.T.H., Parmar C., Grossmann P., Carvalho S., Bussink J., Monshouwer R., Haibe-Kains B., Rietveld D., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. - DOI - PMC - PubMed
-
- Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R.G.P.M., Granton P., Zegers C.M.L., Gillies R., Boellard R., Dekker A., et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

