Pharmacogenomics Variability of Lipid-Lowering Therapies in Familial Hypercholesterolemia
- PMID: 34575654
- PMCID: PMC8468752
- DOI: 10.3390/jpm11090877
Pharmacogenomics Variability of Lipid-Lowering Therapies in Familial Hypercholesterolemia
Abstract
The exponential expansion of genomic data coupled with the lack of appropriate clinical categorization of the variants is posing a major challenge to conventional medications for many common and rare diseases. To narrow this gap and achieve the goals of personalized medicine, a collaborative effort should be made to characterize the genomic variants functionally and clinically with a massive global genomic sequencing of "healthy" subjects from several ethnicities. Familial-based clustered diseases with homogenous genetic backgrounds are amongst the most beneficial tools to help address this challenge. This review will discuss the diagnosis, management, and clinical monitoring of familial hypercholesterolemia patients from a wide angle to cover both the genetic mutations underlying the phenotype, and the pharmacogenomic traits unveiled by the conventional and novel therapeutic approaches. Achieving a drug-related interactive genomic map will potentially benefit populations at risk across the globe who suffer from dyslipidemia.
Keywords: PCSK9 inhibitors; ezetimibe; familial hypercholesterolemia; novel lipid-lowering therapy; pharmacogenomics; statins.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any relevant affiliations or commercial or financial relationships that could be construed as a potential conflict of interest. No writing assistance was used in preparation of the manuscript.
Figures
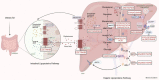

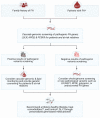
References
-
- Hayat M., Kerr R., Bentley A.R., Rotimi C.N., Raal F.J., Ramsay M. Genetic associations between serum low LDL-cholesterol levels and variants in LDLR, APOB, PCSK9 and LDLRAP1 in African populations. PLoS ONE. 2020;15:e0229098. doi: 10.1371/journal.pone.0249478. Correction in 2021, 16, e0249478. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

