The Cdc14 Phosphatase Controls Resolution of Recombination Intermediates and Crossover Formation during Meiosis
- PMID: 34575966
- PMCID: PMC8470964
- DOI: 10.3390/ijms22189811
The Cdc14 Phosphatase Controls Resolution of Recombination Intermediates and Crossover Formation during Meiosis
Abstract
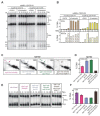
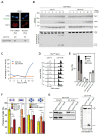
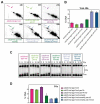
Meiotic defects derived from incorrect DNA repair during gametogenesis can lead to mutations, aneuploidies and infertility. The coordinated resolution of meiotic recombination intermediates is required for crossover formation, ultimately necessary for the accurate completion of both rounds of chromosome segregation. Numerous master kinases orchestrate the correct assembly and activity of the repair machinery. Although much less is known, the reversal of phosphorylation events in meiosis must also be key to coordinate the timing and functionality of repair enzymes. Cdc14 is a crucial phosphatase required for the dephosphorylation of multiple CDK1 targets in many eukaryotes. Mutations that inactivate this phosphatase lead to meiotic failure, but until now it was unknown if Cdc14 plays a direct role in meiotic recombination. Here, we show that the elimination of Cdc14 leads to severe defects in the processing and resolution of recombination intermediates, causing a drastic depletion in crossovers when other repair pathways are compromised. We also show that Cdc14 is required for the correct activity and localization of the Holliday Junction resolvase Yen1/GEN1. We reveal that Cdc14 regulates Yen1 activity from meiosis I onwards, and this function is essential for crossover resolution in the absence of other repair pathways. We also demonstrate that Cdc14 and Yen1 are required to safeguard sister chromatid segregation during the second meiotic division, a late action that is independent of the earlier role in crossover formation. Thus, this work uncovers previously undescribed functions of the evolutionary conserved Cdc14 phosphatase in the regulation of meiotic recombination.
Keywords: CDK1; Cdc14; Cdc20; Cdc5; Holliday junction; Mus81; Ndt80; Sgs1; Yen1; aneuploidy; meiotic recombination.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Zakharyevich K., Ma Y., Tang S., Hwang P.Y., Boiteux S., Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: Double-strand break resection and resolution of double Holliday junctions. Mol. Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. - DOI - PMC - PubMed
-
- Sanchez A., Adam C., Rauh F., Duroc Y., Ranjha L., Lombard B., Mu X., Wintrebert M., Loew D., Guarne A., et al. Exo1 recruits Cdc5 polo kinase to MutLgamma to ensure efficient meiotic crossover formation. Proc. Natl. Acad. Sci. USA. 2020;117:30577–30588. doi: 10.1073/pnas.2013012117. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

