Targeting Autophagy with Natural Products as a Potential Therapeutic Approach for Cancer
- PMID: 34575981
- PMCID: PMC8467030
- DOI: 10.3390/ijms22189807
Targeting Autophagy with Natural Products as a Potential Therapeutic Approach for Cancer
Abstract
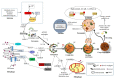
Macro-autophagy (autophagy) is a highly conserved eukaryotic intracellular process of self-digestion caused by lysosomes on demand, which is upregulated as a survival strategy upon exposure to various stressors, such as metabolic insults, cytotoxic drugs, and alcohol abuse. Paradoxically, autophagy dysfunction also contributes to cancer and aging. It is well known that regulating autophagy by targeting specific regulatory molecules in its machinery can modulate multiple disease processes. Therefore, autophagy represents a significant pharmacological target for drug development and therapeutic interventions in various diseases, including cancers. According to the framework of autophagy, the suppression or induction of autophagy can exert therapeutic properties through the promotion of cell death or cell survival, which are the two main events targeted by cancer therapies. Remarkably, natural products have attracted attention in the anticancer drug discovery field, because they are biologically friendly and have potential therapeutic effects. In this review, we summarize the up-to-date knowledge regarding natural products that can modulate autophagy in various cancers. These findings will provide a new position to exploit more natural compounds as potential novel anticancer drugs and will lead to a better understanding of molecular pathways by targeting the various autophagy stages of upcoming cancer therapeutics.
Keywords: anticancer drugs; autophagy; autophagy modulators; mTOR signaling; natural products; resveratrol; ω-3 PUFAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Horibe A., Eid N., Ito Y., Hamaoka H., Tanaka Y., Kondo Y. Upregulated autophagy in Sertoli cells of ethanol-treated rats is associated with induction of inducible nitric oxide synthase (iNOS), androgen receptor suppression and germ cell apoptosis. Int. J. Mol. Sci. 2017;18:1061. doi: 10.3390/ijms18051061. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

