Pharmacological Characterization of Low Molecular Weight Biased Agonists at the Follicle Stimulating Hormone Receptor
- PMID: 34576014
- PMCID: PMC8469697
- DOI: 10.3390/ijms22189850
Pharmacological Characterization of Low Molecular Weight Biased Agonists at the Follicle Stimulating Hormone Receptor
Abstract
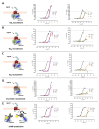
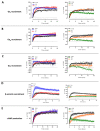
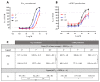
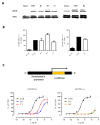
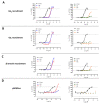
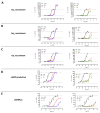
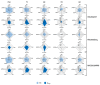
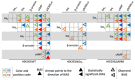
Follicle-stimulating hormone receptor (FSHR) plays a key role in reproduction through the activation of multiple signaling pathways. Low molecular weight (LMW) ligands composed of biased agonist properties are highly valuable tools to decipher complex signaling mechanisms as they allow selective activation of discrete signaling cascades. However, available LMW FSHR ligands have not been fully characterized yet. In this context, we explored the pharmacological diversity of three benzamide and two thiazolidinone derivatives compared to FSH. Concentration/activity curves were generated for Gαs, Gαq, Gαi, β-arrestin 2 recruitment, and cAMP production, using BRET assays in living cells. ERK phosphorylation was analyzed by Western blotting, and CRE-dependent transcription was assessed using a luciferase reporter assay. All assays were done in either wild-type, Gαs or β-arrestin 1/2 CRISPR knockout HEK293 cells. Bias factors were calculated for each pair of read-outs by using the operational model. Our results show that each ligand presented a discrete pharmacological efficacy compared to FSH, ranging from super-agonist for β-arrestin 2 recruitment to pure Gαs bias. Interestingly, LMW ligands generated kinetic profiles distinct from FSH (i.e., faster, slower or transient, depending on the ligand) and correlated with CRE-dependent transcription. In addition, clear system biases were observed in cells depleted of either Gαs or β-arrestin genes. Such LMW properties are useful pharmacological tools to better dissect the multiple signaling pathways activated by FSHR and assess their relative contributions at the cellular and physio-pathological levels.
Keywords: FSHR; allosteric ligands; biased signaling; operational model; system bias.
Conflict of interest statement
S.G.N., H.N.Y. and S.S.P. were founders and employees of TocopheRx Inc.
Figures

References
MeSH terms
Substances
Grants and funding
- ANR-10-LABX-53; ARD2020 Biomédicaments/Agence Nationale de la Recherche
- APR-IR MODUPHAC/Conseil Régional du Centre-Val de Loire
- 665790/H2020 Marie Skłodowska-Curie Actions
- PRIME 18gm5910013; LEAP 18gm0010004/Japan Agency for Medical Research and Development
- KAKENHI 17K08264/Japan Society for the Promotion of Science
LinkOut - more resources
Full Text Sources
Miscellaneous

