Quinolone Resistance of Actinobacillus pleuropneumoniae Revealed through Genome and Transcriptome Analyses
- PMID: 34576206
- PMCID: PMC8472844
- DOI: 10.3390/ijms221810036
Quinolone Resistance of Actinobacillus pleuropneumoniae Revealed through Genome and Transcriptome Analyses
Abstract
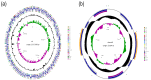
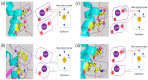
Actinobacillus pleuropneumoniae is a pathogen that infects pigs and poses a serious threat to the pig industry. The emergence of quinolone-resistant strains of A.pleuropneumoniae further limits the choice of treatment. However, the mechanisms behind quinolone resistance in A.pleuropneumoniae remain unclear. The genomes of a ciprofloxacin-resistant strain, A. pleuropneumoniae SC1810 and its isogenic drug-sensitive counterpart were sequenced and analyzed using various bioinformatics tools, revealing 559 differentially expressed genes. The biological membrane, plasmid-mediated quinolone resistance genes and quinolone resistance-determining region were detected. Upregulated expression of efflux pump genes led to ciprofloxacin resistance. The expression of two porins, OmpP2B and LamB, was significantly downregulated in the mutant. Three nonsynonymous mutations in the mutant strain disrupted the water-metal ion bridge, subsequently reducing the affinity of the quinolone-enzyme complex for metal ions and leading to cross-resistance to multiple quinolones. The mechanism of quinolone resistance in A. pleuropneumoniae may involve inhibition of expression of the outer membrane protein genes ompP2B and lamB to decrease drug influx, overexpression of AcrB in the efflux pump to enhance its drug-pumping ability, and mutation in the quinolone resistance-determining region to weaken the binding of the remaining drugs. These findings will provide new potential targets for treatment.
Keywords: Actinobacillus pleuropneumoniae; efflux pump; porin; quinolone resistance; quinolone resistance-determining region; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Stringer O.W., Bosse J.T., Lacouture S., Gottschalk M., Fodor L., Angen O., Velazquez E., Penny P., Lei L., Langford P.R., et al. Proposal of Actinobacillus pleuropneumoniae serovar 19, and reformulation of previous multiplex PCRs for capsule-specific typing of all known serovars. Vet. Microbiol. 2021;255:109021. doi: 10.1016/j.vetmic.2021.109021. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

