Phytate and Microbial Suspension Amendments Increased Soybean Growth and Shifted Microbial Community Structure
- PMID: 34576699
- PMCID: PMC8471086
- DOI: 10.3390/microorganisms9091803
Phytate and Microbial Suspension Amendments Increased Soybean Growth and Shifted Microbial Community Structure
Abstract
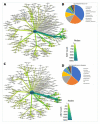
Phytate represents an organic pool of phosphorus in soil that requires hydrolysis by phytase enzymes produced by microorganisms prior to its bioavailability by plants. We tested the ability of a microbial suspension made from an old growth maple forest's undisturbed soil to mineralize phytate in a greenhouse trial on soybean plants inoculated or non-inoculated with the suspension. MiSeq Amplicon sequencing targeting bacterial 16S rRNA gene and fungal ITS was performed to assess microbial community changes following treatments. Our results showed that soybean nodulation and shoot dry weight biomass increased when phytate was applied to the nutrient-poor substrate mixture. Bacterial and fungal diversities of the root and rhizosphere biotopes were relatively resilient following inoculation by microbial suspension; however, bacterial community structure was significantly influenced. Interestingly, four arbuscular mycorrhizal fungi (AMF) were identified as indicator species, including Glomus sp., Claroideoglomus etunicatum, Funneliformis mosseae and an unidentified AMF taxon. We also observed that an ericoid mycorrhizal taxon Sebacina sp. and three Trichoderma spp. were among indicator species. Non-pathogenic Planctobacteria members highly dominated the bacterial community as core and hub taxa for over 80% of all bacterial datasets in root and rhizosphere biotopes. Overall, our study documented that inoculation with a microbial suspension and phytate amendment improved soybean plant growth.
Keywords: MiSeq; microbiome; network; phosphorus; phytate; soybean.
Conflict of interest statement
There is no conflict of interests among the authors.
Figures





References
-
- FAO . World Fertilizer Trends and Outlook to 2020: Summary Report. FAO; Rome, Italy: 2017.
-
- Obersteiner M., Peñuelas J., Ciais P., van der Velde M., Janssens I.A. The phosphorus trilemma. Nat. Geosci. 2013;6:897–898. doi: 10.1038/ngeo1990. - DOI
-
- Jarosch K.A., Kandeler E., Frossard E., Bünemann E. Is the enzymatic hydrolysis of soil organic phosphorus compounds limited by enzyme or substrate availability? Soil Biol. Biochem. 2019;139:107628. doi: 10.1016/j.soilbio.2019.107628. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

