Glucosylation of (±)-Menthol by Uridine-Diphosphate-Sugar Dependent Glucosyltransferases from Plants
- PMID: 34576983
- PMCID: PMC8470988
- DOI: 10.3390/molecules26185511
Glucosylation of (±)-Menthol by Uridine-Diphosphate-Sugar Dependent Glucosyltransferases from Plants
Abstract
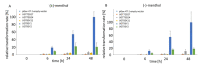
Menthol is a cyclic monoterpene alcohol of the essential oils of plants of the genus Mentha, which is in demand by various industries due to its diverse sensorial and physiological properties. However, its poor water solubility and its toxic effect limit possible applications. Glycosylation offers a solution as the binding of a sugar residue to small molecules increases their water solubility and stability, renders aroma components odorless and modifies bioactivity. In order to identify plant enzymes that catalyze this reaction, a glycosyltransferase library containing 57 uridine diphosphate sugar-dependent enzymes (UGTs) was screened with (±)-menthol. The identity of the products was confirmed by mass spectrometry and nuclear magnetic resonance spectroscopy. Five enzymes were able to form (±)-menthyl-β-d-glucopyranoside in whole-cell biotransformations: UGT93Y1, UGT93Y2, UGT85K11, UGT72B27 and UGT73B24. In vitro enzyme activity assays revealed highest catalytic activity for UGT93Y1 (7.6 nkat/mg) from Camellia sinensis towards menthol and its isomeric forms. Although UGT93Y2 shares 70% sequence identity with UGT93Y1, it was less efficient. Of the five enzymes, UGT93Y1 stood out because of its high in vivo and in vitro biotransformation rate. The identification of novel menthol glycosyltransferases from the tea plant opens new perspectives for the biotechnological production of menthyl glucoside.
Keywords: Camellia sinensis; UDP-glucosyltransferase; UGT93Y1; menthol; menthyl glucoside.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Härtner J., Reinscheid U.M. Conformational analysis of menthol diastereomers by NMR and DFT computation. J. Mol. Struct. 2008;872:145–149. doi: 10.1016/j.molstruc.2007.02.029. - DOI
-
- Fahlbusch K.-G., Hammerschmidt F.-J., Panten J., Pickenhagen W., Schatkowski D., Bauer K., Garbe D., Surburg H. Ullmann’s Encyclopedia of Industrial Chemistry. Wiley Online Library; Hoboken, NJ, USA: 2005. Flavors and Fragrances. - DOI
-
- Jerković I., Mastelić J. Composition of free and glycosidically bound volatiles of Mentha aquatica L. Croat. Chem. Acta. 2001;74:431–439.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

