Semaphorin 3A-Glycosaminoglycans Interaction as Therapeutic Target for Axonal Regeneration
- PMID: 34577606
- PMCID: PMC8465649
- DOI: 10.3390/ph14090906
Semaphorin 3A-Glycosaminoglycans Interaction as Therapeutic Target for Axonal Regeneration
Abstract
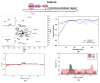
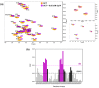
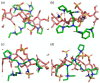
Semaphorin 3A (Sema3A) is a cell-secreted protein that participates in the axonal guidance pathways. Sema3A acts as a canonical repulsive axon guidance molecule, inhibiting CNS regenerative axonal growth and propagation. Therefore, interfering with Sema3A signaling is proposed as a therapeutic target for achieving functional recovery after CNS injuries. It has been shown that Sema3A adheres to the proteoglycan component of the extracellular matrix (ECM) and selectively binds to heparin and chondroitin sulfate-E (CS-E) glycosaminoglycans (GAGs). We hypothesize that the biologically relevant interaction between Sema3A and GAGs takes place at Sema3A C-terminal polybasic region (SCT). The aims of this study were to characterize the interaction of the whole Sema3A C-terminal polybasic region (Sema3A 725-771) with GAGs and to investigate the disruption of this interaction by small molecules. Recombinant Sema3A basic domain was produced and we used a combination of biophysical techniques (NMR, SPR, and heparin affinity chromatography) to gain insight into the interaction of the Sema3A C-terminal domain with GAGs. The results demonstrate that SCT is an intrinsically disordered region, which confirms that SCT binds to GAGs and helps to identify the specific residues involved in the interaction. NMR studies, supported by molecular dynamics simulations, show that a new peptoid molecule (CSIC02) may disrupt the interaction between SCT and heparin. Our structural study paves the way toward the design of new molecules targeting these protein-GAG interactions with potential therapeutic applications.
Keywords: NMR; glycosaminoglycan–protein interaction; peptoids; semaphorin 3A.
Conflict of interest statement
Some of the authors (Messeguer, À.; Alfonso, I.; Bujons, J.; Pérez, Y.; Corredor, M.; Moure, A.) are co-inventors of a patent application including CSIC02 and its biological activity against Sema3A (see Section 5). The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

