Production of Thermophilic Chitinase by Paenibacillus sp. TKU052 by Bioprocessing of Chitinous Fishery Wastes and Its Application in N-acetyl-D-glucosamine Production
- PMID: 34577952
- PMCID: PMC8471714
- DOI: 10.3390/polym13183048
Production of Thermophilic Chitinase by Paenibacillus sp. TKU052 by Bioprocessing of Chitinous Fishery Wastes and Its Application in N-acetyl-D-glucosamine Production
Abstract
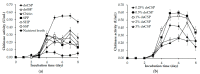
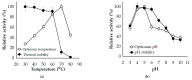
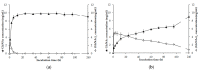
The bioprocessing of chitinous fishery wastes (CFWs) to chitinases through fermentation approaches has gained importance owing to its great benefits in reducing the enzyme production cost, and utilizing chitin waste. In this work, our study of the chitinase production of Paenibacillus sp. TKU052 in the presence of different kinds of CFWs revealed a preference for demineralized crab shells powder (deCSP); furthermore, a 72 kDa chitinase was isolated from the 0.5% deCSP-containing medium. The Paenibacillus sp. TKU052 chitinase displayed maximum activity at 70 °C and pH 4-5, while Zn2+, Fe3+, Triton X-100, Tween 40, and SDS exerted a negative effect on its activity, whereas Mn2+ and 2-mercaptoethanol were found to potentially enhance the activity. Among various kinds of polysaccharide, Paenibacillus sp. TKU052 chitinase exhibited the best catalytic activity on colloidal chitin (CC) with Km = 9.75 mg/mL and Vmax = 2.43 μmol/min. The assessment of the hydrolysis of CC and N-acetyl chitooligosaccharides revealed that Paenibacillus sp. TKU052 chitinase possesses multiple catalytic functions, including exochitinase, endochitinase, and N-acetyl-β-D-glucosaminidase activities. Finally, the combination of Paenibacillus sp. TKU052 chitinase and Streptomyces speibonae TKU048 N-acetyl-β-D-glucosaminidase could efficiently convert CC to N-acetyl-D-glucosamine (GlcNAc) with a production yield of 94.35-98.60% in 12-24 h.
Keywords: N-acetyl-D-glucosamine; Paenibacillus; chitinase; chitinous fishery wastes; crab shells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
An Exochitinase with N-Acetyl-β-Glucosaminidase-Like Activity from Shrimp Head Conversion by Streptomyces speibonae and Its Application in Hydrolyzing β-Chitin Powder to Produce N-Acetyl-d-Glucosamine.Polymers (Basel). 2019 Sep 30;11(10):1600. doi: 10.3390/polym11101600. Polymers (Basel). 2019. PMID: 31574975 Free PMC article.
-
An acidic, thermostable exochitinase with β-N-acetylglucosaminidase activity from Paenibacillus barengoltzii converting chitin to N-acetyl glucosamine.Biotechnol Biofuels. 2014 Dec 10;7(1):174. doi: 10.1186/s13068-014-0174-y. eCollection 2014. Biotechnol Biofuels. 2014. PMID: 25550712 Free PMC article.
-
Utilization of Seafood Processing By-Products for Production of Proteases by Paenibacillus sp. TKU052 and Their Application in Biopeptides' Preparation.Mar Drugs. 2020 Nov 20;18(11):574. doi: 10.3390/md18110574. Mar Drugs. 2020. PMID: 33233577 Free PMC article.
-
Purification and characterization of chitinase from Paenibacillus sp. .Biotechnol Appl Biochem. 2021 Feb;68(1):30-40. doi: 10.1002/bab.1889. Epub 2020 Feb 19. Biotechnol Appl Biochem. 2021. PMID: 31957084 Review.
-
Structures and functions of carbohydrate-active enzymes of chitinolytic bacteria Paenibacillus sp. str. FPU-7.Biosci Biotechnol Biochem. 2021 May 25;85(6):1314-1323. doi: 10.1093/bbb/zbab058. Biosci Biotechnol Biochem. 2021. PMID: 33792636 Review.
Cited by
-
Effects of Modified Glucosamine on the Chondrogenic Potential of Circulating Stem Cells under Experimental Inflammation.Int J Mol Sci. 2023 Jun 20;24(12):10397. doi: 10.3390/ijms241210397. Int J Mol Sci. 2023. PMID: 37373540 Free PMC article.
-
The Impact of Chitinase Binding Domain Truncation on the Properties of CaChi18B from Chitinilyticum aquatile CSC-1.Mar Drugs. 2025 Feb 20;23(3):93. doi: 10.3390/md23030093. Mar Drugs. 2025. PMID: 40137279 Free PMC article.
-
Chitosanase Production from the Liquid Fermentation of Squid Pens Waste by Paenibacillus elgii.Polymers (Basel). 2023 Sep 11;15(18):3724. doi: 10.3390/polym15183724. Polymers (Basel). 2023. PMID: 37765578 Free PMC article.
-
Production, Purification, and Characterization of a Cellulase from Paenibacillus elgii.Polymers (Basel). 2024 Jul 17;16(14):2037. doi: 10.3390/polym16142037. Polymers (Basel). 2024. PMID: 39065354 Free PMC article.
-
Enzymes from Fishery and Aquaculture Waste: Research Trends in the Era of Artificial Intelligence and Circular Bio-Economy.Mar Drugs. 2024 Sep 10;22(9):411. doi: 10.3390/md22090411. Mar Drugs. 2024. PMID: 39330292 Free PMC article. Review.
References
-
- Yadav M., Goswami P., Paritosh K., Kumar M., Pareek N., Vivekanand V. Seafood waste: A source for preparation of commercially employable chitin/chitosan materials. Bioresources. 2019;6:8–28. doi: 10.1186/s40643-019-0243-y. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

