Oxidative Stress Response in Pseudomonas aeruginosa
- PMID: 34578219
- PMCID: PMC8466533
- DOI: 10.3390/pathogens10091187
Oxidative Stress Response in Pseudomonas aeruginosa
Abstract
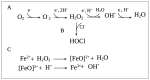
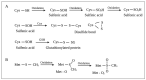
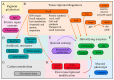
Pseudomonas aeruginosa is a Gram-negative environmental and human opportunistic pathogen highly adapted to many different environmental conditions. It can cause a wide range of serious infections, including wounds, lungs, the urinary tract, and systemic infections. The high versatility and pathogenicity of this bacterium is attributed to its genomic complexity, the expression of several virulence factors, and its intrinsic resistance to various antimicrobials. However, to thrive and establish infection, P. aeruginosa must overcome several barriers. One of these barriers is the presence of oxidizing agents (e.g., hydrogen peroxide, superoxide, and hypochlorous acid) produced by the host immune system or that are commonly used as disinfectants in a variety of different environments including hospitals. These agents damage several cellular molecules and can cause cell death. Therefore, bacteria adapt to these harsh conditions by altering gene expression and eliciting several stress responses to survive under oxidative stress. Here, we used PubMed to evaluate the current knowledge on the oxidative stress responses adopted by P. aeruginosa. We will describe the genes that are often differently expressed under oxidative stress conditions, the pathways and proteins employed to sense and respond to oxidative stress, and how these changes in gene expression influence pathogenicity and the virulence of P. aeruginosa. Understanding these responses and changes in gene expression is critical to controlling bacterial pathogenicity and developing new therapeutic agents.
Keywords: Pseudomonas aeruginosa; antimicrobial resistance; bacterial stress response; oxidative stress; oxidative stress response; reactive chlorine species; reactive oxygen species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Heimesaat M.M., Escher U., Grunau A., Kühl A.A., Bereswill S. Multidrug-Resistant Pseudomonas aeruginosa Accelerate Intestinal, Extra-Intestinal, and Systemic Inflammatory Responses in Human Microbiota-Associated Mice With Subacute Ileitis. Front. Immunol. 2019;10:49. doi: 10.3389/fimmu.2019.00049. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

