Tracing Viral Transmission and Evolution of Bovine Leukemia Virus through Long Read Oxford Nanopore Sequencing of the Proviral Genome
- PMID: 34578223
- PMCID: PMC8470207
- DOI: 10.3390/pathogens10091191
Tracing Viral Transmission and Evolution of Bovine Leukemia Virus through Long Read Oxford Nanopore Sequencing of the Proviral Genome
Abstract
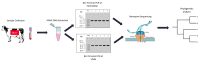
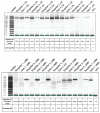
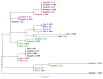
Bovine leukemia virus (BLV) causes Enzootic Bovine Leukosis (EBL), a persistent life-long disease resulting in immune dysfunction and shortened lifespan in infected cattle, severely impacting the profitability of the US dairy industry. Our group has found that 94% of dairy farms in the United States are infected with BLV with an average in-herd prevalence of 46%. This is partly due to the lack of clinical presentation during the early stages of primary infection and the elusive nature of BLV transmission. This study sought to validate a near-complete genomic sequencing approach for reliability and accuracy before determining its efficacy in characterizing the sequence identity of BLV proviral genomes collected from a pilot study made up of 14 animals from one commercial dairy herd. These BLV-infected animals were comprised of seven adult dam/daughter pairs that tested positive by ELISA and qPCR. The results demonstrate sequence identity or divergence of the BLV genome from the same samples tested in two independent laboratories, suggesting both vertical and horizontal transmission in this dairy herd. This study supports the use of Oxford Nanopore sequencing for the identification of viral SNPs that can be used for retrospective genetic contact tracing of BLV transmission.
Keywords: Oxford Nanopore Sequencing; bovine leukemia virus; phylogenetics; proviral load; retroviral evolution; targeted sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bartlett P., Michigan State University Bovine Leukemia Virus in the U.S.: Impact and Options for Control #1 What Is It? Why Does It Matter? What Options for Control? Where to Start? What Can USAHA Do? [(accessed on 14 September 2021)]. Available online: https://blv.msu.edu/resources/USAHA-Oct-16-2017.pdf.
LinkOut - more resources
Full Text Sources

