Evaluation of Inhibitory Activity In Silico of In-House Thiomorpholine Compounds between the ACE2 Receptor and S1 Subunit of SARS-CoV-2 Spike
- PMID: 34578240
- PMCID: PMC8468748
- DOI: 10.3390/pathogens10091208
Evaluation of Inhibitory Activity In Silico of In-House Thiomorpholine Compounds between the ACE2 Receptor and S1 Subunit of SARS-CoV-2 Spike
Abstract
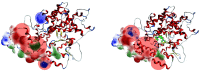
At the end of 2019, the world was struck by the COVID-19 pandemic, which resulted in dire repercussions of unimaginable proportions. From the beginning, the international scientific community employed several strategies to tackle the spread of this disease. Most notably, these consisted of the development of a COVID-19 vaccine and the discovery of antiviral agents through the repositioning of already known drugs with methods such as de novo design. Previously, methylthiomorphic compounds, designed by our group as antihypertensive agents, have been shown to display an affinity with the ACE2 (angiotensin converting enzyme) receptor, a key mechanism required for SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) entry into target cells. Therefore, the objective of this work consists of evaluating, in silico, the inhibitory activity of these compounds between the ACE2 receptor and the S1 subunit of the SARS-CoV-2 spike protein. Supported by the advances of different research groups on the structure of the coronavirus spike and the interaction of the latter with its receptor, ACE2, we carried out a computational study that examined the effect of in-house designed compounds on the inhibition of said interaction. Our results indicate that the polyphenol LQM322 is one of the candidates that should be considered as a possible anti-COVID-19 agent.
Keywords: SARS-CoV-2; antivirals; spike; thiomorpholine derivatives.
Conflict of interest statement
The authors declare no conflict of interest.
Figures












References
-
- World Health Organization WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19. 11 March 2020. [(accessed on 18 July 2021)]. Available online: https://www.who.int/director-general/speeches/detail/who-director-genera....
-
- World Health Organization Weekly Epidemiological Update on COVID-19—13 July 2021. [(accessed on 18 July 2021)]. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on....
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

