SARS-CoV-2 Infection: New Molecular, Phylogenetic, and Pathogenetic Insights. Efficacy of Current Vaccines and the Potential Risk of Variants
- PMID: 34578269
- PMCID: PMC8473168
- DOI: 10.3390/v13091687
SARS-CoV-2 Infection: New Molecular, Phylogenetic, and Pathogenetic Insights. Efficacy of Current Vaccines and the Potential Risk of Variants
Abstract
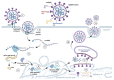
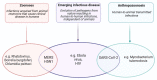
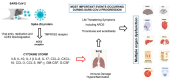
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a newly discovered coronavirus responsible for the coronavirus disease 2019 (COVID-19) pandemic. COVID-19 has rapidly become a public health emergency of international concern. Although remarkable scientific achievements have been reached since the beginning of the pandemic, the knowledge behind this novel coronavirus, in terms of molecular and pathogenic characteristics and zoonotic potential, is still relatively limited. Today, there is a vaccine, or rather several vaccines, which, for the first time in the history of highly contagious infectious diseases that have plagued mankind, has been manufactured in just one year. Currently, four vaccines are licensed by regulatory agencies, and they use RNA or viral vector technologies. The positive effects of the vaccination campaign are being felt in many parts of the world, but the disappearance of this new infection is still far from being a reality, as it is also threatened by the presence of novel SARS-CoV-2 variants that could undermine the effectiveness of the vaccine, hampering the immunization control efforts. Indeed, the current findings indicate that SARS-CoV-2 is adapting to transmission in humans more efficiently, while further divergence from the initial archetype should be considered. In this review, we aimed to provide a collection of the current knowledge regarding the molecular, phylogenetic, and pathogenetic insights into SARS-CoV-2. The most recent findings obtained with respect to the impact of novel emerging SARS-CoV-2 variants as well as the development and implementation of vaccines are highlighted.
Keywords: COVID-19; CoV; SARS-CoV-2; Severe acute respiratory syndrome coronavirus 2; coronavirus disease 2019; coronaviruses; spike protein; vaccine; variants; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M., et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavi. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

