Genomic Characterisation of a Highly Divergent Siadenovirus (Psittacine Siadenovirus F) from the Critically Endangered Orange-Bellied Parrot (Neophema chrysogaster)
- PMID: 34578295
- PMCID: PMC8472863
- DOI: 10.3390/v13091714
Genomic Characterisation of a Highly Divergent Siadenovirus (Psittacine Siadenovirus F) from the Critically Endangered Orange-Bellied Parrot (Neophema chrysogaster)
Abstract
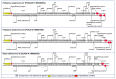
Siadenoviruses have been detected in wild and captive birds worldwide. Only nine siadenoviruses have been fully sequenced; however, partial sequences for 30 others, many of these from wild Australian birds, are also described. Some siadenoviruses, e.g., the turkey siadenovirus A, can cause disease; however, most cause subclinical infections. An example of a siadenovirus causing predominately subclinical infections is psittacine siadenovirus 2, proposed name psittacine siadenovirus F (PsSiAdV-F), which is enzootic in the captive breeding population of the critically endangered orange-bellied parrot (OBP, Neophema chrysogaster). Here, we have fully characterised PsSiAdV-F from an OBP. The PsSiAdV-F genome is 25,392 bp in length and contained 25 putative genes. The genome architecture of PsSiAdV-F exhibited characteristics similar to members within the genus Siadenovirus; however, the novel PsSiAdV-F genome was highly divergent, showing highest and lowest sequence similarity to skua siadenovirus A (57.1%) and psittacine siadenovirus D (31.1%), respectively. Subsequent phylogenetic analyses of the novel PsSiAdV-F genome positioned the virus into a phylogenetically distinct sub-clade with all other siadenoviruses and did not show any obvious close evolutionary relationship. Importantly, the resulted tress continually demonstrated that novel PsSiAdV-F evolved prior to all known members except the frog siadenovirus A in the evolution and possibly the ancestor of the avian siadenoviruses. To date, PsSiAdV-F has not been detected in wild parrots, so further studies screening PsSiAdV-F in wild Australian parrots and generating whole genome sequences of siadenoviruses of Australian native passerine species is recommended to fill the siadenovirus evolutionary gaps.
Keywords: Adenoviridae; evolution; next-generation sequencing; orange-bellied parrot; psittacine siadenovirus F; siadenovirus.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- ICTV Virus Taxonomy—2020 Release. [(accessed on 18 August 2021)]. Available online: https://talk.ictvonline.org/taxonomy/
-
- Rivera S., Wellehan J., McManamon R., Innis C., Garner M., Raphael B., Gregory C., Latimer K., Rodriguez C., Figueroa O., et al. Systemic adenovirus infection in Sulawesi tortoises (Indotestudo forsteni) caused by a novel siadenovirus. J. Vet. Diagn. Invest. 2009;21:415–426. doi: 10.1177/104063870902100402. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

