Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions
- PMID: 34578318
- PMCID: PMC8472933
- DOI: 10.3390/v13091737
Structural Analysis of the Menangle Virus P Protein Reveals a Soft Boundary between Ordered and Disordered Regions
Abstract
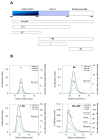
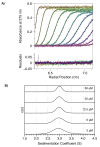
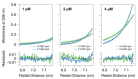
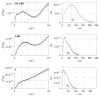
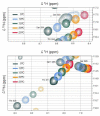
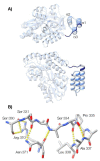
The paramyxoviral phosphoprotein (P protein) is the non-catalytic subunit of the viral RNA polymerase, and coordinates many of the molecular interactions required for RNA synthesis. All paramyxoviral P proteins oligomerize via a centrally located coiled-coil that is connected to a downstream binding domain by a dynamic linker. The C-terminal region of the P protein coordinates interactions between the catalytic subunit of the polymerase, and the viral nucleocapsid housing the genomic RNA. The inherent flexibility of the linker is believed to facilitate polymerase translocation. Here we report biophysical and structural characterization of the C-terminal region of the P protein from Menangle virus (MenV), a bat-borne paramyxovirus with zoonotic potential. The MenV P protein is tetrameric but can dissociate into dimers at sub-micromolar protein concentrations. The linker is globally disordered and can be modeled effectively as a worm-like chain. However, NMR analysis suggests very weak local preferences for alpha-helical and extended beta conformation exist within the linker. At the interface between the disordered linker and the structured C-terminal binding domain, a gradual disorder-to-order transition occurs, with X-ray crystallographic analysis revealing a dynamic interfacial structure that wraps the surface of the binding domain.
Keywords: Paramyxoviruses; RNA-dependent RNA polymerase; X-ray crystallography; analytical ultracentrifugation; conformational exchange; intrinsically disordered proteins; pararubulaviruses; protein self-association; small-angle X-ray scattering; solution NMR spectroscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

