Zika Virus Non-Structural Protein 1 Antigen-Capture Immunoassay
- PMID: 34578352
- PMCID: PMC8473068
- DOI: 10.3390/v13091771
Zika Virus Non-Structural Protein 1 Antigen-Capture Immunoassay
Abstract
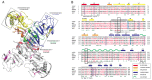
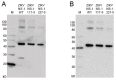
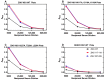
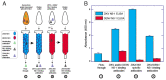
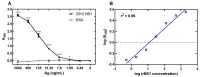
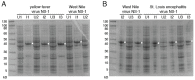
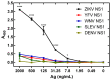
Infection with Zika virus (ZIKV), a member of the Flavivirus genus of the Flaviviridae family, typically results in mild self-limited illness, but severe neurological disease occurs in a limited subset of patients. In contrast, serious outcomes commonly occur in pregnancy that affect the developing fetus, including microcephaly and other major birth defects. The genetic similarity of ZIKV to other widespread flaviviruses, such as dengue virus (DENV), presents a challenge to the development of specific ZIKV diagnostic assays. Nonstructural protein 1 (NS1) is established for use in immunodiagnostic assays for flaviviruses. To address the cross-reactivity of ZIKV NS1 with proteins from other flaviviruses we used site-directed mutagenesis to modify putative epitopes. Goat polyclonal antibodies to variant ZIKV NS1 were affinity-purified to remove antibodies binding to the closely related NS1 protein of DENV. An antigen-capture ELISA configured with the affinity-purified polyclonal antibody showed a linear dynamic range between approximately 500 and 30 ng/mL, with a limit of detection of between 1.95 and 7.8 ng/mL. NS1 proteins from DENV, yellow fever virus, St. Louis encephalitis virus and West Nile virus showed significantly reduced reactivity in the ZIKV antigen-capture ELISA. Refinement of approaches similar to those employed here could lead to development of ZIKV-specific immunoassays suitable for use in areas where infections with related flaviviruses are common.
Keywords: Zika virus; antigen-capture ELISA; non-structural protein 1; polyclonal antibodies; site-directed mutagenesis.
Conflict of interest statement
R.F.G. is a co-founder of Zalgen Labs, a biotechnology company developing countermeasures for emerging viruses. All other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- Smithburen K.C., Kerr J.A., Gatne P.B. Neutralizing antibodies against certain viruses in the sera of residents of India. J. Immunol. 1954;72:248–257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

