RNA Structures and Their Role in Selective Genome Packaging
- PMID: 34578369
- PMCID: PMC8472981
- DOI: 10.3390/v13091788
RNA Structures and Their Role in Selective Genome Packaging
Abstract
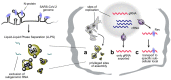
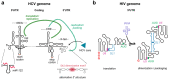
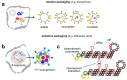
To generate infectious viral particles, viruses must specifically select their genomic RNA from milieu that contains a complex mixture of cellular or non-genomic viral RNAs. In this review, we focus on the role of viral encoded RNA structures in genome packaging. We first discuss how packaging signals are constructed from local and long-range base pairings within viral genomes, as well as inter-molecular interactions between viral and host RNAs. Then, how genome packaging is regulated by the biophysical properties of RNA. Finally, we examine the impact of RNA packaging signals on viral evolution.
Keywords: RNA; RNA structure; RNA virus; evolution; genome packaging; viral assembly.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the writing of the manuscript, or in the decision to publish.
Figures



References
-
- Wilkinson K., Gorelick R.J., Vasa S.M., Guex N., Rein A., Mathews D.H., Giddings M.C., Weeks K.M. High-Throughput SHAPE Analysis Reveals Structures in HIV-1 Genomic RNA Strongly Conserved across Distinct Biological States. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

