A Bioluminescent 3CLPro Activity Assay to Monitor SARS-CoV-2 Replication and Identify Inhibitors
- PMID: 34578395
- PMCID: PMC8473059
- DOI: 10.3390/v13091814
A Bioluminescent 3CLPro Activity Assay to Monitor SARS-CoV-2 Replication and Identify Inhibitors
Abstract
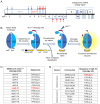
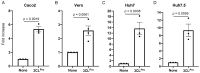
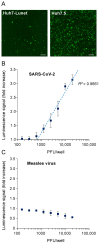
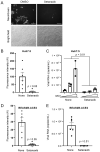
Our therapeutic arsenal against viruses is very limited and the current pandemic of SARS-CoV-2 highlights the critical need for effective antivirals against emerging coronaviruses. Cellular assays allowing a precise quantification of viral replication in high-throughput experimental settings are essential to the screening of chemical libraries and the selection of best antiviral chemical structures. To develop a reporting system for SARS-CoV-2 infection, we generated cell lines expressing a firefly luciferase maintained in an inactive form by a consensus cleavage site for the viral protease 3CLPro of coronaviruses, so that the luminescent biosensor is turned on upon 3CLPro expression or SARS-CoV-2 infection. This cellular assay was used to screen a metabolism-oriented library of 492 compounds to identify metabolic vulnerabilities of coronaviruses for developing innovative therapeutic strategies. In agreement with recent reports, inhibitors of pyrimidine biosynthesis were found to prevent SARS-CoV-2 replication. Among the top hits, we also identified the NADPH oxidase (NOX) inhibitor Setanaxib. The anti-SARS-CoV-2 activity of Setanaxib was further confirmed using ACE2-expressing human pulmonary cells Beas2B as well as human primary nasal epithelial cells. Altogether, these results validate our cell-based functional assay and the interest of screening libraries of different origins to identify inhibitors of SARS-CoV-2 for drug repurposing or development.
Keywords: BAY2402234; DHODH; IPPA-17-A04; NADPH oxidase; SARS-CoV-2; Setanaxib; Vidofludimus; antiviral; chemical screening.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Ahmed-Belkacem R., Sutto-Ortiz P., Guiraud M., Canard B., Vasseur J.-J., Decroly E., Debart F. Synthesis of Adenine Dinucleosides SAM Analogs as Specific Inhibitors of SARS-CoV Nsp14 RNA Cap Guanine-N7-Methyltransferase. Eur. J. Med. Chem. 2020;201:112557. doi: 10.1016/j.ejmech.2020.112557. - DOI - PMC - PubMed
-
- Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 Antiviral Drugs through Large-Scale Compound Repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

