Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte
- PMID: 34578418
- PMCID: PMC8473176
- DOI: 10.3390/v13091837
Investigating the Viral Suppressor HC-Pro Inhibiting Small RNA Methylation through Functional Comparison of HEN1 in Angiosperm and Bryophyte
Abstract
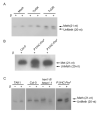
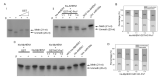
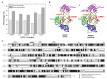
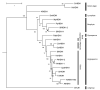
In plants, HEN1-facilitated methylation at 3' end ribose is a critical step of small-RNA (sRNA) biogenesis. A mutant of well-studied Arabidopsis HEN1 (AtHEN1), hen1-1, showed a defective developmental phenotype, indicating the importance of sRNA methylation. Moreover, Marchantia polymorpha has been identified to have a HEN1 ortholog gene (MpHEN1); however, its function remained unfathomed. Our in vivo and in vitro data have shown MpHEN1 activity being comparable with AtHEN1, and their substrate specificity towards duplex microRNA (miRNA) remained consistent. Furthermore, the phylogenetic tree and multiple alignment highlighted the conserved molecular evolution of the HEN1 family in plants. The P1/HC-Pro of the turnip mosaic virus (TuMV) is a known RNA silencing suppressor and inhibits HEN1 methylation of sRNAs. Here, we report that the HC-Pro physically binds with AtHEN1 through FRNK motif, inhibiting HEN1's methylation activity. Moreover, the in vitro EMSA data indicates GST-HC-Pro of TuMV lacks sRNA duplex-binding ability. Surprisingly, the HC-Pro also inhibits MpHEN1 activity in a dosage-dependent manner, suggesting the possibility of interaction between HC-Pro and MpHEN1 as well. Further investigations on understanding interaction mechanisms of HEN1 and various HC-Pros can advance the knowledge of viral suppressors.
Keywords: HEN1; HEN1-HC-Pro interaction; methylation; sRNA; viral suppressors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

