Frontiers in the Standardization of the Plant Platform for High Scale Production of Vaccines
- PMID: 34579360
- PMCID: PMC8467261
- DOI: 10.3390/plants10091828
Frontiers in the Standardization of the Plant Platform for High Scale Production of Vaccines
Abstract
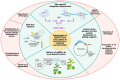
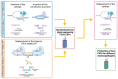
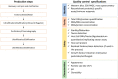
The recent COVID-19 pandemic has highlighted the value of technologies that allow a fast setup and production of biopharmaceuticals in emergency situations. The plant factory system can provide a fast response to epidemics/pandemics. Thanks to their scalability and genome plasticity, plants represent advantageous platforms to produce vaccines. Plant systems imply less complicated production processes and quality controls with respect to mammalian and bacterial cells. The expression of vaccines in plants is based on transient or stable transformation systems and the recent progresses in genome editing techniques, based on the CRISPR/Cas method, allow the manipulation of DNA in an efficient, fast, and easy way by introducing specific modifications in specific sites of a genome. Nonetheless, CRISPR/Cas is far away from being fully exploited for vaccine expression in plants. In this review, an overview of the potential conjugation of the renewed vaccine technologies (i.e., virus-like particles-VLPs, and industrialization of the production process) with genome editing to produce vaccines in plants is reported, illustrating the potential advantages in the standardization of the plant platforms, with the overtaking of constancy of large-scale production challenges, facilitating regulatory requirements and expediting the release and commercialization of the vaccine products of genome edited plants.
Keywords: genome editing; plant factory system; vaccines; virus-like particles.
Conflict of interest statement
C.B., C.C. and L.C. declare no conflict of interest. F.C. is an employee of the GSK group of companies. GSK Vaccines Institute for Global Health Srl is an affiliate of GlaxoSmithKline Biologicals SA.
Figures



References
-
- Ward B.J., Makarkov A., Seguin A., Pillet S., Trepanier S., Dhaliwall J., Libman M.D., Vesikari T., Landry N. Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (>/=65 years): Two multicentre, randomised phase 3 trials. Lancet. 2020;396:1491–1503. doi: 10.1016/S0140-6736(20)32014-6. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

