The Arabinogalactan Protein Family of Centaurium erythraea Rafn
- PMID: 34579403
- PMCID: PMC8471777
- DOI: 10.3390/plants10091870
The Arabinogalactan Protein Family of Centaurium erythraea Rafn
Abstract
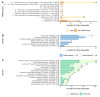
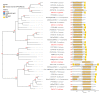
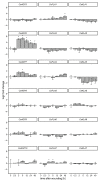
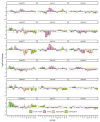
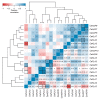
Centaurium erythraea (centaury) is a medicinal plant with exceptional developmental plasticity in vitro and vigorous, often spontaneous, regeneration via shoot organogenesis and somatic embryogenesis, during which arabinogalactan proteins (AGPs) play an important role. AGPs are highly glycosylated proteins belonging to the super family of O-glycosylated plant cell surface hydroxyproline-rich glycoproteins (HRGPs). HRGPs/AGPs are intrinsically disordered and not well conserved, making their homology-based mining ineffective. We have applied a recently developed pipeline for HRGP/AGP mining, ragp, which is based on machine learning prediction of proline hydroxylation, to identify HRGP sequences in centaury transcriptome and to classify them into motif and amino acid bias (MAAB) classes. AGP sequences with low AG glycomotif representation were also identified. Six members of each of the three AGP subclasses, fasciclin-like AGPs, receptor kinase-like AGPs and AG peptides, were selected for phylogenetic and expression analyses. The expression of these 18 genes was recorded over 48 h following leaf mechanical wounding, as well as in 16 tissue samples representing plants from nature, plants cultivated in vitro, and developmental stages during shoot organogenesis and somatic embryogenesis. None of the selected genes were upregulated during both wounding recovery and regeneration. Possible functions of AGPs with the most interesting expression profiles are discussed.
Keywords: FLAs; MAAB classes; arabinogalactan proteins; gene expression; hydroxyproline-rich glycoproteins; mechanical wounding; organogenesis; protein kinases; ragp; somatic embryogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Deepak S., Shailasree S., Kini R.K., Muck A., Mithöfer A., Shetty S.H. Hydroxyproline-rich Glycoproteins and Plant Defence. J. Phytopathol. 2010;158:585–593. doi: 10.1111/j.1439-0434.2010.01669.x. - DOI
-
- Hijazi M., Velasquez S.M., Jamet E., Estevez J.M., Albenne C. An update on post-translational modifications of hydroxyproline-rich glycoproteins: Toward a model highlighting their contribution to plant cell wall architecture. Front. Plant Sci. 2014;5:395. doi: 10.3389/fpls.2014.00395. - DOI - PMC - PubMed
-
- Kieliszewski M.J., Lamport D.T.A., Tan L., Cannon M.C. Hydroxyproline-Rich Glycoproteins: Form and Function. Annu. Plant Rev. 2010;41:321–342. doi: 10.1002/9781444391015.ch13. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

