SARS-CoV-2 Mutations and their Viral Variants
- PMID: 34580015
- PMCID: PMC8252702
- DOI: 10.1016/j.cytogfr.2021.06.001
SARS-CoV-2 Mutations and their Viral Variants
Abstract
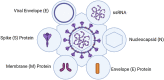
Mutations in the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) occur spontaneously during replication. Thousands of mutations have accumulated and continue to since the emergence of the virus. As novel mutations continue appearing at the scene, naturally, new variants are increasingly observed. Since the first occurrence of the SARS-CoV-2 infection, a wide variety of drug compounds affecting the binding sites of the virus have begun to be studied. As the drug and vaccine trials are continuing, it is of utmost importance to take into consideration the SARS-CoV-2 mutations and their respective frequencies since these data could lead the way to multi-drug combinations. The lack of effective therapeutic and preventive strategies against human coronaviruses (hCoVs) necessitates research that is of interest to the clinical applications. The reason why the mutations in glycoprotein S lead to vaccine escape is related to the location of the mutation and the affinity of the protein. At the same time, it can be said that variations should occur in areas such as the receptor-binding domain (RBD), and vaccines and antiviral drugs should be formulated by targeting more than one viral protein. In this review, a literature survey in the scope of the increasing SARS-CoV-2 mutations and the viral variations is conducted. In the light of current knowledge, the various disguises of the mutant SARS-CoV-2 forms and their apparent differences from the original strain are examined as they could possibly aid in finding the most appropriate therapeutic approaches.
Keywords: COVID-19; Mutation; Receptor-binding domain; SARS-CoV-2; Spike protein; Viral variants.
Copyright © 2021. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare there are no competing interests.
Figures


References
-
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R., Walls A.C., Kemp S.A., Bassi J., Pinto D., Fregni C.S., Bianchi S., Tortorici M.A., Bowen J., Culap K., Jaconi S., Cameroni E., Snell G., Pizzuto M.S., Pellanda A.F., Garzoni C., Riva A., Elmer A., Kingston N., Graves B., Mccoy L.E., Smith K.G., Bradley J.R., Thaventhiran J., Ceron-Gutierrez L., Barcenas-Morales G., Virgin H.W., Lanzavecchia A., Piccoli L., Doffinger R., Wills M., Veesler D., Corti D., Gupta R.K. SARS-CoV-2 B.1.1.7 escape from mRNA vaccine-elicited neutralizing antibodies The CITIID-NIHR BioResource COVID-19 Collaboration. MedRxiv. 2021 doi: 10.1101/2021.01.19.21249840. 2021.01.19.21249840. - DOI - PubMed
-
- Finkel Y., Mizrahi O., Nachshon A., Weingarten-Gabbay S., Morgenstern D., Yahalom-Ronen Y., Tamir H., Achdout H., Stein D., Israeli O., Beth-Din A., Melamed S., Weiss S., Israely T., Paran N., Schwartz M., Stern-Ginossar N. The coding capacity of SARS-CoV-2. Nature. 2021;589:125–130. doi: 10.1038/s41586-020-2739-1. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

