Genetic variants associated with platelet count are predictive of human disease and physiological markers
- PMID: 34580418
- PMCID: PMC8476563
- DOI: 10.1038/s42003-021-02642-9
Genetic variants associated with platelet count are predictive of human disease and physiological markers
Abstract
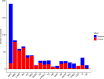
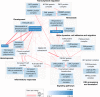
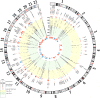
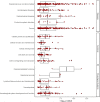
Platelets play an important role in hemostasis and other aspects of vascular biology. We conducted a meta-analysis of platelet count GWAS using data on 536,974 Europeans and identified 577 independent associations. To search for mechanisms through which these variants affect platelets, we applied cis-expression quantitative trait locus, DEPICT and IPA analyses and assessed genetic sharing between platelet count and various traits using polygenic risk scoring. We found genetic sharing between platelet count and counts of other blood cells (except red blood cells), in addition to several other quantitative traits, including markers of cardiovascular, liver and kidney functions, height, and weight. Platelet count polygenic risk score was predictive of myeloproliferative neoplasms, rheumatoid arthritis, ankylosing spondylitis, hypertension, and benign prostate hyperplasia. Taken together, these results advance understanding of diverse aspects of platelet biology and how they affect biological processes in health and disease.
© 2021. The Author(s).
Conflict of interest statement
E. Mikaelsdottir, G. Thorleifsson, L. Stefansdottir, G. Halldorsson, J.K. Sigurdsson, S.H. Lund, V. Tragante, P. Melsted, S. Rognvaldsson, K. Norland, A. Helgadottir, M. K. Magnusson, J. Gudmundsson, H. Holm, S. Saevarsdottir, I. Jonsdottir, G. Thorgeirsson, D.F. Gudbjartsson, U. Thorsteinsdottir, T. Rafnar, and K. Stefansson, who are affiliated with deCODE genetics/Amgen, declare competing financial interests as employees. The remaining authors declare no competing financial interests. There are no non-financial competing interests to declare.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

